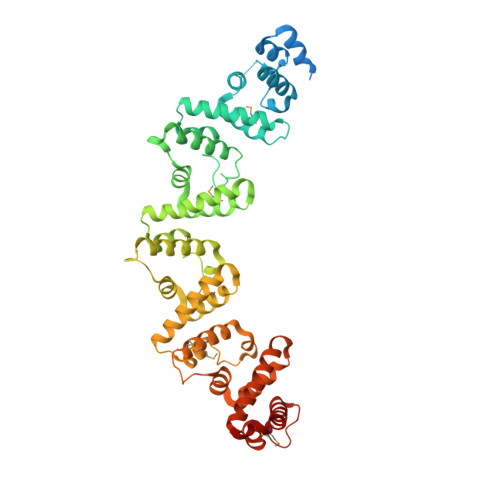
Crystal structure of the RxLR effector PcRxLR12 from Phytophthora capsici
Zhao, L., Zhang, X., Zhang, X., Song, W., Li, X., Feng, R., Yang, C., Huang, Z., Zhu, C.(2018) Biochem Biophys Res Commun 503: 1830-1835
- PubMed: 30077372
- DOI: https://doi.org/10.1016/j.bbrc.2018.07.121
- Primary Citation of Related Structures:
5ZC3 - PubMed Abstract:
RxLR genes are a prominent class of effectors in oomycetes, and almost half of these proteins contain a conserved sequence motif termed the WY domain, that may exist singly, or as divergent tandem repeats in different effectors. Here we describe the crystal structure of PcRxLR12 (63-488) from Phytophthora capsici at 3.0 Å resolution. The structure consists of five tandemly arrayed WY-domains linked to each other by short connecting helices. Superposition of the WY-2 domain on the other four domains of PcRxLR12, show that the first α-helix termed the K motif, and Loop 3 which connects α 3 and α 4 are the key regions of structural divergence between the WY domains. A similar pattern was observed when WY-2 was superposed on the 11 WY domains from other oomycete effectors. We also note that an added connecting helix between WY domains in some RXLR effectors, ensures that the WY domains are oriented in the same direction.
- College of Plant Protection, Shandong Agricultural University, Tai'an, China; Shandong Provincial Key Laboratory for Biology of Vegetable Diseases and Insect, Shandong Agricultural University, Tai'an, China.
Organizational Affiliation: