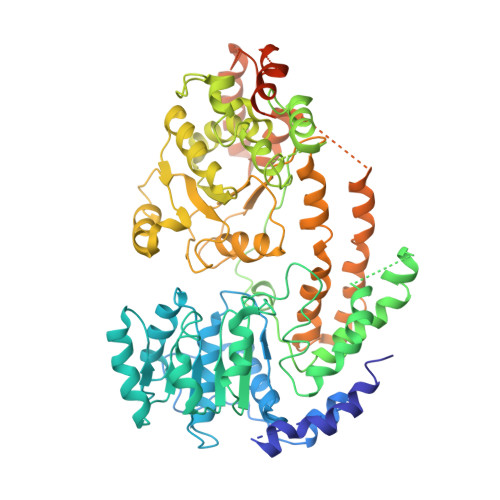
Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Li, M., Xia, X., Tian, Y., Jia, Q., Liu, X., Lu, Y., Li, M., Li, X., Chen, Z.(2019) Nature 567: 409-413
- PubMed: 30867599
- DOI: https://doi.org/10.1038/s41586-019-1029-2
- Primary Citation of Related Structures:
5Z3L, 5Z3O, 5Z3U, 5Z3V, 6IY2, 6IY3 - PubMed Abstract:
Chromatin remodellers include diverse enzymes with distinct biological functions, but nucleosome-sliding activity appears to be a common theme 1,2 . Among the remodelling enzymes, Snf2 serves as the prototype to study the action of this protein family. Snf2 and related enzymes share two conserved RecA-like lobes 3 , which by themselves are able to couple ATP hydrolysis to chromatin remodelling. The mechanism by which these enzymes couple ATP hydrolysis to translocate the nucleosome along the DNA remains unclear 2,4-8 . Here we report the structures of Saccharomyces cerevisiae Snf2 bound to the nucleosome in the presence of ADP and ADP-BeF x . Snf2 in the ADP-bound state adopts an open conformation similar to that in the apo state, and induces a one-base-pair DNA bulge at superhelix location 2 (SHL2), with the tracking strand showing greater distortion than the guide strand. The DNA distortion propagates to the proximal end, leading to staggered translocation of the two strands. The binding of ADP-BeF x triggers a closed conformation of the enzyme, resetting the nucleosome to a relaxed state. Snf2 shows altered interactions with the DNA in different nucleotide states, providing the structural basis for DNA translocation. Together, our findings suggest a fundamental mechanism for the DNA translocation that underlies chromatin remodelling.
- MOE Key Laboratory of Protein Science, Tsinghua University, Beijing, China.
Organizational Affiliation: