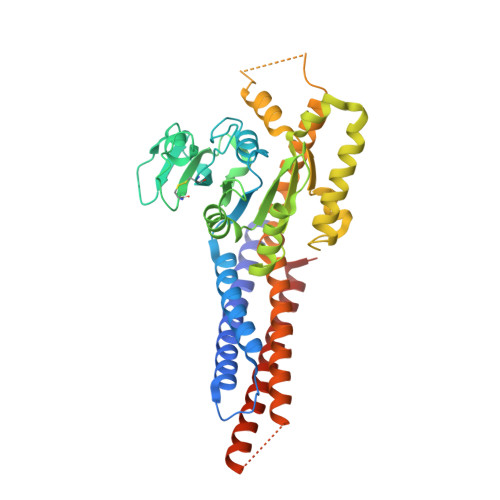
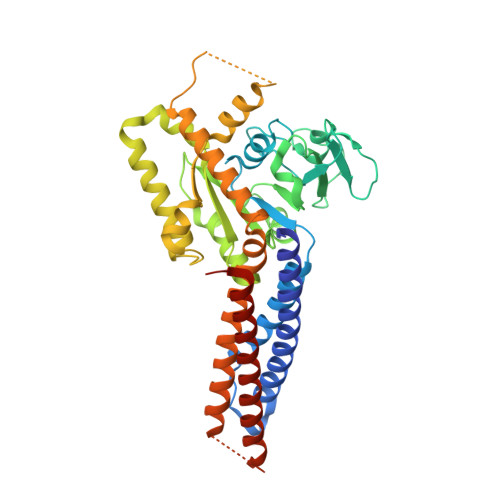
Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion
Yan, L., Qi, Y., Huang, X., Yu, C., Lan, L., Guo, X., Rao, Z., Hu, J., Lou, Z.(2018) Nat Struct Mol Biol 25: 233-243
- PubMed: 29483649
- DOI: https://doi.org/10.1038/s41594-018-0034-8
- Primary Citation of Related Structures:
5YEW - PubMed Abstract:
Fusion of the outer mitochondrial membrane is mediated by the dynamin-like GTPase mitofusin (MFN). Here, we determined the structure of the minimal GTPase domain (MGD) of human MFN1 in complex with GDP-BeF 3 - . The MGD folds into a canonical GTPase fold with an associating four-helix bundle, HB1, and forms a dimer. A potassium ion in the catalytic core engages GDP and BeF 3 - (GDP-BeF 3 - ). Enzymatic analysis has confirmed that efficient GTP hydrolysis by MFN1 requires potassium. Compared to previously reported MGD structures, the HB1 structure undergoes a major conformational change relative to the GTPase domains, as they move from pointing in opposite directions to point in the same direction, suggesting that a swing of the four-helix bundle can pull tethered membranes closer to achieve fusion. The proposed model is supported by results from in vitro biochemical assays and mitochondria morphology rescue assays in MFN1-deleted cells. These findings offer an explanation for how Charcot-Marie-Tooth neuropathy type 2 A (CMT2A)-causing mutations compromise MFN-mediated fusion.
- School of Medicine, Tsinghua University, Beijing, China.
Organizational Affiliation: