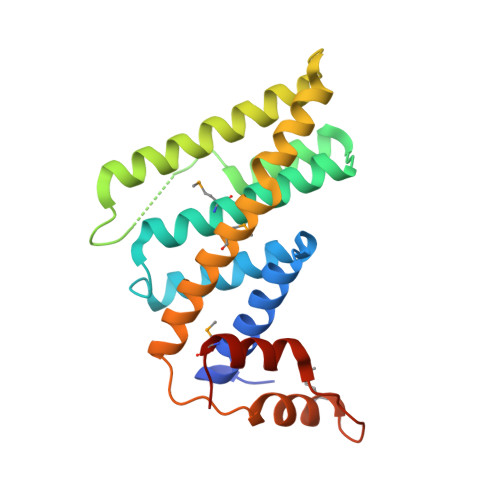
Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex.
Hu, C., Rai, R., Huang, C., Broton, C., Long, J., Xu, Y., Xue, J., Lei, M., Chang, S., Chen, Y.(2017) Cell Res 27: 1485-1502
- PubMed: 29160297
- DOI: https://doi.org/10.1038/cr.2017.144
- Primary Citation of Related Structures:
5XYF - PubMed Abstract:
Telomeres are nucleoprotein complexes that play essential roles in protecting chromosome ends. Mammalian telomeres consist of repetitive DNA sequences bound by the shelterin complex. In this complex, the POT1-TPP1 heterodimer binds to single-stranded telomeric DNAs, while TRF1 and TRF2-RAP1 interact with double-stranded telomeric DNAs. TIN2, the linchpin of this complex, simultaneously interacts with TRF1, TRF2, and TPP1 to mediate the stable assembly of the shelterin complex. However, the molecular mechanism by which TIN2 interacts with these proteins to orchestrate telomere protection remains poorly understood. Here, we report the crystal structure of the N-terminal domain of TIN2 in complex with TIN2-binding motifs from TPP1 and TRF2, revealing how TIN2 interacts cooperatively with TPP1 and TRF2. Unexpectedly, TIN2 contains a telomeric repeat factor homology (TRFH)-like domain that functions as a protein-protein interaction platform. Structure-based mutagenesis analyses suggest that TIN2 plays an important role in maintaining the stable shelterin complex required for proper telomere end protection.
- State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Science Research Center, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences; University of Chinese Academy of Sciences, 333 Haike Road, Shanghai 201210, China.
Organizational Affiliation: