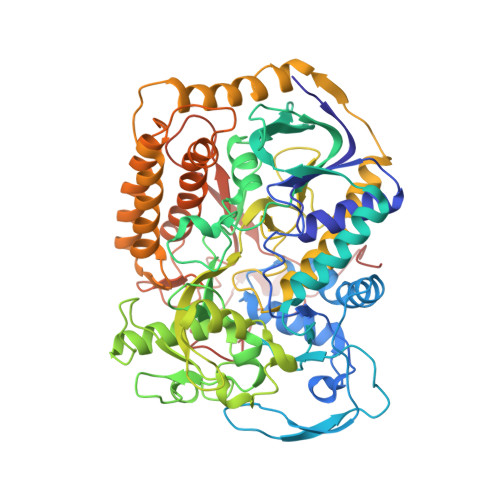
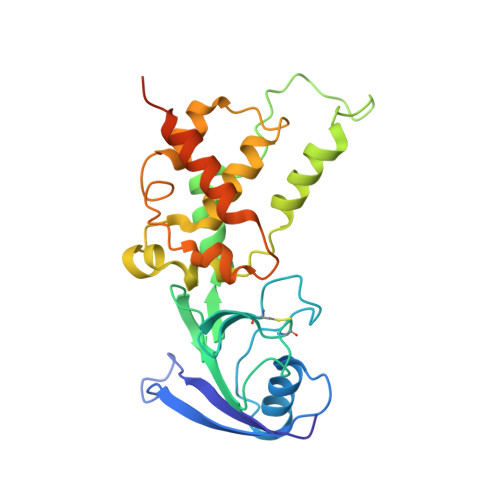
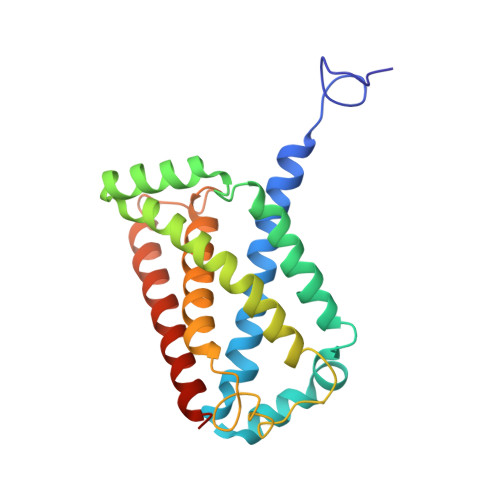
Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas.
Guan, H.H., Hsieh, Y.C., Lin, P.J., Huang, Y.C., Yoshimura, M., Chen, L.Y., Chen, S.K., Chuankhayan, P., Lin, C.C., Chen, N.C., Nakagawa, A., Chan, S.I., Chen, C.J.(2018) Sci Rep 8: 14935-14935
- PubMed: 30297797
- DOI: https://doi.org/10.1038/s41598-018-33193-5
- Primary Citation of Related Structures:
5XMJ - PubMed Abstract:
The membrane-embedded quinol:fumarate reductase (QFR) in anaerobic bacteria catalyzes the reduction of fumarate to succinate by quinol in the anaerobic respiratory chain. The electron/proton-transfer pathways in QFRs remain controversial. Here we report the crystal structure of QFR from the anaerobic sulphate-reducing bacterium Desulfovibrio gigas (D. gigas) at 3.6 Å resolution. The structure of the D. gigas QFR is a homo-dimer, each protomer comprising two hydrophilic subunits, A and B, and one transmembrane subunit C, together with six redox cofactors including two b-hemes. One menaquinone molecule is bound near heme b L in the hydrophobic subunit C. This location of the menaquinone-binding site differs from the menaquinol-binding cavity proposed previously for QFR from Wolinella succinogenes. The observed bound menaquinone might serve as an additional redox cofactor to mediate the proton-coupled electron transport across the membrane. Armed with these structural insights, we propose electron/proton-transfer pathways in the quinol reduction of fumarate to succinate in the D. gigas QFR.
- Life Science Group, Scientific Research Division, National Synchrotron Radiation Research Center, Hsinchu, 30076, Taiwan.
Organizational Affiliation: