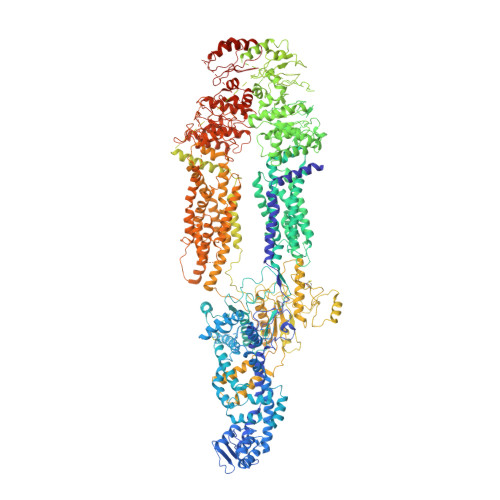
Structure of the Human Lipid Exporter ABCA1.
Qian, H., Zhao, X., Cao, P., Lei, J., Yan, N., Gong, X.(2017) Cell 169: 1228-1239.e10
- PubMed: 28602350
- DOI: https://doi.org/10.1016/j.cell.2017.05.020
- Primary Citation of Related Structures:
5XJY - PubMed Abstract:
ABCA1, an ATP-binding cassette (ABC) subfamily A exporter, mediates the cellular efflux of phospholipids and cholesterol to the extracellular acceptor apolipoprotein A-I (apoA-I) for generation of nascent high-density lipoprotein (HDL). Mutations of human ABCA1 are associated with Tangier disease and familial HDL deficiency. Here, we report the cryo-EM structure of human ABCA1 with nominal resolutions of 4.1 Å for the overall structure and 3.9 Å for the massive extracellular domain. The nucleotide-binding domains (NBDs) display a nucleotide-free state, while the two transmembrane domains (TMDs) contact each other through a narrow interface in the intracellular leaflet of the membrane. In addition to TMDs and NBDs, two extracellular domains of ABCA1 enclose an elongated hydrophobic tunnel. Structural mapping of dozens of disease-related mutations allows potential interpretation of their diverse pathogenic mechanisms. Structural-based analysis suggests a plausible "lateral access" mechanism for ABCA1-mediated lipid export that may be distinct from the conventional alternating-access paradigm.
- State Key Laboratory of Membrane Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China; Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: