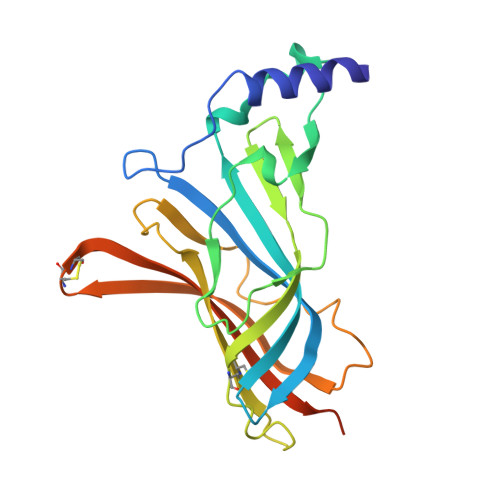
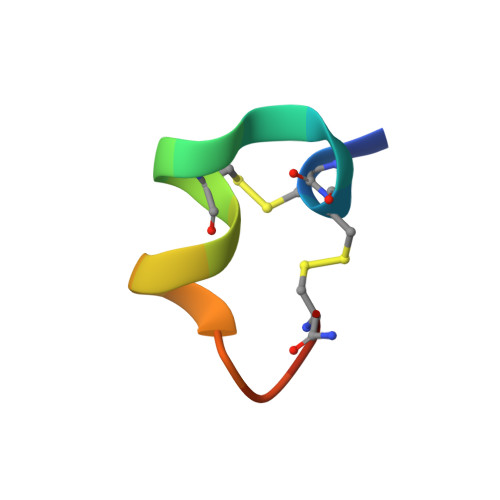
The crystal structure of Ac-AChBP in complex with alpha-conotoxin LvIA reveals the mechanism of its selectivity towards different nAChR subtypes
Xu, M., Zhu, X., Yu, J., Yu, J., Luo, S., Wang, X.(2017) Protein Cell 8: 675-685
- PubMed: 28585176
- DOI: https://doi.org/10.1007/s13238-017-0426-2
- Primary Citation of Related Structures:
5XGL - PubMed Abstract:
The α3* nAChRs, which are considered to be promising drug targets for problems such as pain, addiction, cardiovascular function, cognitive disorders etc., are found throughout the central and peripheral nervous system. The α-conotoxin (α-CTx) LvIA has been identified as the most selective inhibitor of α3β2 nAChRs known to date, and it can distinguish the α3β2 nAChR subtype from the α6/α3β2β3 and α3β4 nAChR subtypes. However, the mechanism of its selectivity towards α3β2, α6/α3β2β3, and α3β4 nAChRs remains elusive. Here we report the co-crystal structure of LvIA in complex with Aplysia californica acetylcholine binding protein (Ac-AChBP) at a resolution of 3.4 Å. Based on the structure of this complex, together with homology modeling based on other nAChR subtypes and binding affinity assays, we conclude that Asp-11 of LvIA plays an important role in the selectivity of LvIA towards α3β2 and α3/α6β2β3 nAChRs by making a salt bridge with Lys-155 of the rat α3 subunit. Asn-9 lies within a hydrophobic pocket that is formed by Met-36, Thr-59, and Phe-119 of the rat β2 subunit in the α3β2 nAChR model, revealing the reason for its more potent selectivity towards the α3β2 nAChR subtype. These results provide molecular insights that can be used to design ligands that selectively target α3β2 nAChRs, with significant implications for the design of new therapeutic α-CTxs.
- The Ministry of Education Key Laboratory of Protein Science, School of Life Sciences, Beijing Advanced Innovation Center for Structural Biology, Collaborative Innovation Center for Biotherapy, Tsinghua University, Beijing, 100084, China.
Organizational Affiliation: