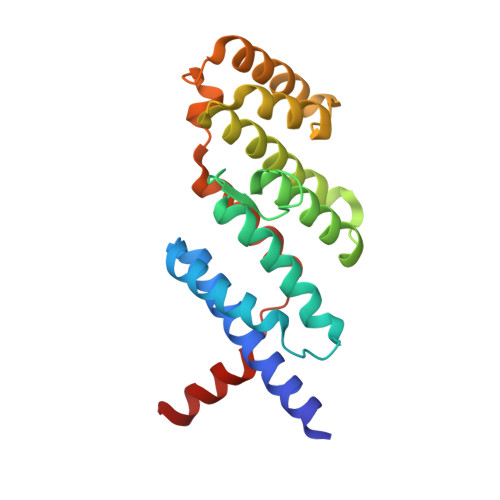
NBS1 Phosphorylation Status Dictates Repair Choice of Dysfunctional Telomeres
Rai, R., Hu, C., Broton, C., Chen, Y., Lei, M., Chang, S.(2017) Mol Cell 65: 801-817.e4
- PubMed: 28216226
- DOI: https://doi.org/10.1016/j.molcel.2017.01.016
- Primary Citation of Related Structures:
5WQD - PubMed Abstract:
Telomeres employ TRF2 to protect chromosome ends from activating the DNA damage sensor MRE11-RAD50-NBS1 (MRN), thereby repressing ATM-dependent DNA damage checkpoint responses. How TRF2 prevents MRN activation at dysfunctional telomeres is unclear. Here, we show that the phosphorylation status of NBS1 determines the repair pathway choice of dysfunctional telomeres. The crystal structure of the TRF2-NBS1 complex at 3.0 Å resolution shows that the NBS1 429 YQLSP 433 motif interacts specifically with the TRF2 TRFH domain. Phosphorylation of NBS1 serine 432 by CDK2 in S/G2 dissociates NBS1 from TRF2, promoting TRF2-Apollo/SNM1B complex formation and the protection of leading-strand telomeres. Classical-NHEJ-mediated repair of telomeres lacking TRF2 requires phosphorylated NBS1 S432 to activate ATM, while interaction of de-phosphorylated NBS1 S432 with TRF2 promotes alternative-NHEJ repair of telomeres lacking POT1-TPP1. Our work advances understanding of how the TRF2 TRFH domain orchestrates telomere end protection and reveals how the phosphorylation status of the NBS1 S432 dictates repair pathway choice of dysfunctional telomeres.
- Department of Laboratory Medicine, Yale University School of Medicine, 330 Cedar Street, New Haven, CT 06520, USA.
Organizational Affiliation: