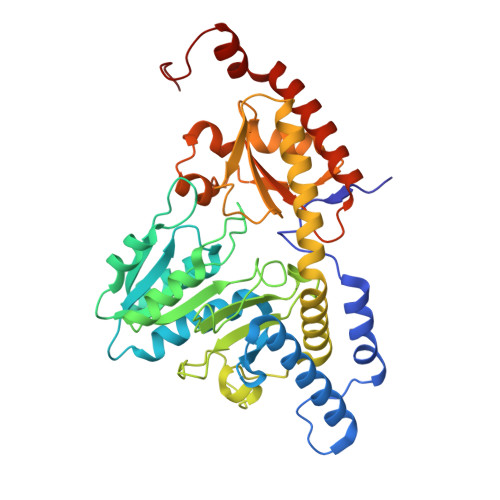
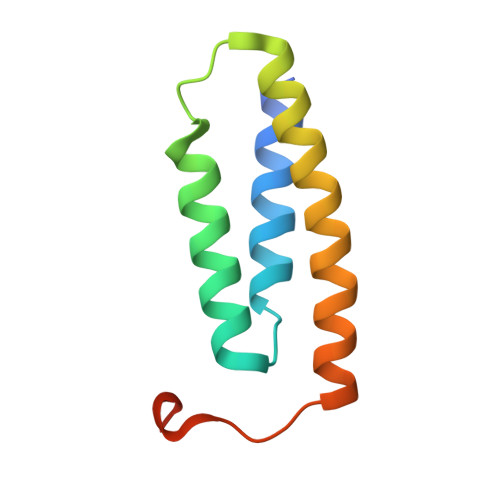
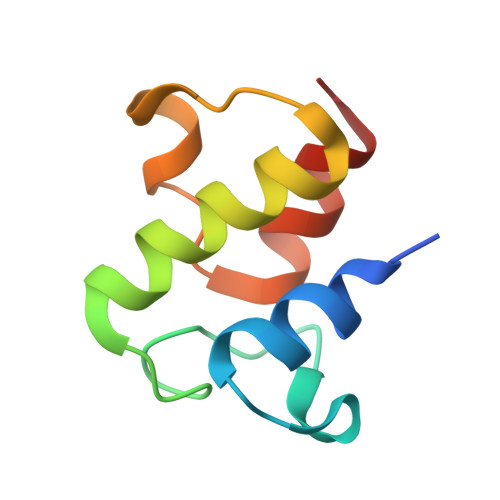
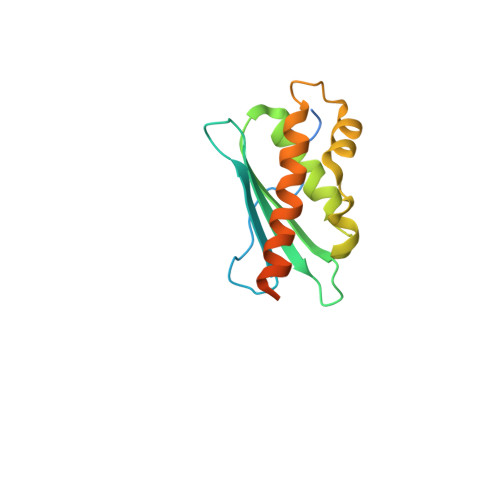
Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex.
Boniecki, M.T., Freibert, S.A., Muhlenhoff, U., Lill, R., Cygler, M.(2017) Nat Commun 8: 1287-1287
- PubMed: 29097656
- DOI: https://doi.org/10.1038/s41467-017-01497-1
- Primary Citation of Related Structures:
5WGB, 5WKP, 5WLW - PubMed Abstract:
Iron-sulfur (Fe/S) clusters are essential protein cofactors crucial for many cellular functions including DNA maintenance, protein translation, and energy conversion. De novo Fe/S cluster synthesis occurs on the mitochondrial scaffold protein ISCU and requires cysteine desulfurase NFS1, ferredoxin, frataxin, and the small factors ISD11 and ACP (acyl carrier protein). Both the mechanism of Fe/S cluster synthesis and function of ISD11-ACP are poorly understood. Here, we present crystal structures of three different NFS1-ISD11-ACP complexes with and without ISCU, and we use SAXS analyses to define the 3D architecture of the complete mitochondrial Fe/S cluster biosynthetic complex. Our structural and biochemical studies provide mechanistic insights into Fe/S cluster synthesis at the catalytic center defined by the active-site Cys of NFS1 and conserved Cys, Asp, and His residues of ISCU. We assign specific regulatory rather than catalytic roles to ISD11-ACP that link Fe/S cluster synthesis with mitochondrial lipid synthesis and cellular energy status.
- Department of Biochemistry, University of Saskatchewan, 107 Wiggins Road, Saskatoon, SK, Canada, S7N 5E5.
Organizational Affiliation: