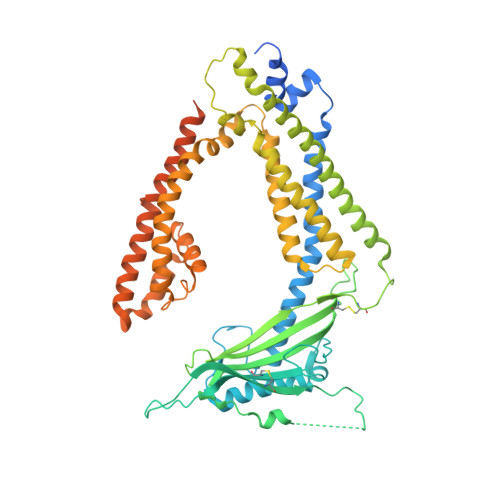
Human TRPML1 channel structures in open and closed conformations.
Schmiege, P., Fine, M., Blobel, G., Li, X.(2017) Nature 550: 366-370
- PubMed: 29019983
- DOI: https://doi.org/10.1038/nature24036
- Primary Citation of Related Structures:
5WJ5, 5WJ9 - PubMed Abstract:
Transient receptor potential mucolipin 1 (TRPML1) is a Ca 2+ -releasing cation channel that mediates the calcium signalling and homeostasis of lysosomes. Mutations in TRPML1 lead to mucolipidosis type IV, a severe lysosomal storage disorder. Here we report two electron cryo-microscopy structures of full-length human TRPML1: a 3.72-Å apo structure at pH 7.0 in the closed state, and a 3.49-Å agonist-bound structure at pH 6.0 in an open state. Several aromatic and hydrophobic residues in pore helix 1, helices S5 and S6, and helix S6 of a neighbouring subunit, form a hydrophobic cavity to house the agonist, suggesting a distinct agonist-binding site from that found in TRPV1, a TRP channel from a different subfamily. The opening of TRPML1 is associated with distinct dilations of its lower gate together with a slight structural movement of pore helix 1. Our work reveals the regulatory mechanism of TRPML channels, facilitates better understanding of TRP channel activation, and provides insights into the molecular basis of mucolipidosis type IV pathogenesis.
- Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, New York, New York 10065, USA.
Organizational Affiliation: