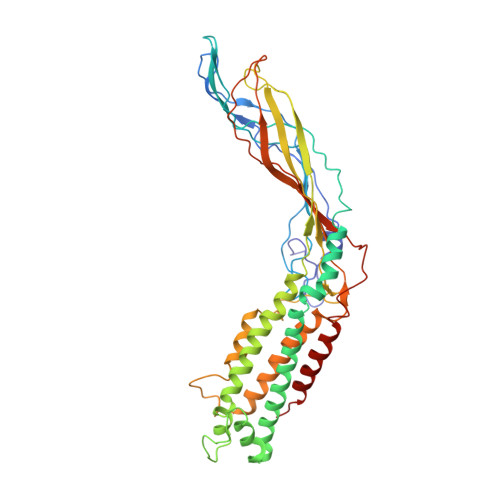
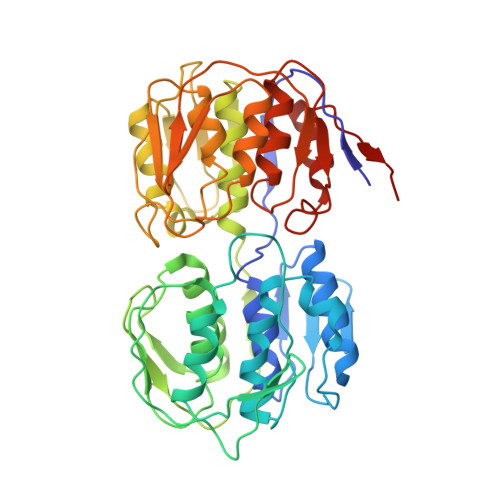
Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Cui, Z., Gorzelnik, K.V., Chang, J.Y., Langlais, C., Jakana, J., Young, R., Zhang, J.(2017) Proc Natl Acad Sci U S A 114: 11697-11702
- PubMed: 29078304
- DOI: https://doi.org/10.1073/pnas.1707102114
- Primary Citation of Related Structures:
5VLY, 5VLZ, 5VM7 - PubMed Abstract:
In single-stranded RNA bacteriophages (ssRNA phages) a single copy of the maturation protein binds the genomic RNA (gRNA) and is required for attachment of the phage to the host pilus. For the canonical Allolevivirus Qβ the maturation protein, A 2 , has an additional role as the lysis protein, by its ability to bind and inhibit MurA, which is involved in peptidoglycan biosynthesis. Here, we determined structures of Qβ virions, virus-like particles, and the Qβ-MurA complex using single-particle cryoelectron microscopy, at 4.7-Å, 3.3-Å, and 6.1-Å resolutions, respectively. We identified the outer surface of the β-region in A 2 as the MurA-binding interface. Moreover, the pattern of MurA mutations that block Qβ lysis and the conformational changes of MurA that facilitate A 2 binding were found to be due to the intimate fit between A 2 and the region encompassing the closed catalytic cleft of substrate-liganded MurA. Additionally, by comparing the Qβ virion with Qβ virus-like particles that lack a maturation protein, we observed a structural rearrangement in the capsid coat proteins that is required to package the viral gRNA in its dominant conformation. Unexpectedly, we found a coat protein dimer sequestered in the interior of the virion. This coat protein dimer binds to the gRNA and interacts with the buried α-region of A 2 , suggesting that it is sequestered during the early stage of capsid formation to promote the gRNA condensation required for genome packaging. These internalized coat proteins are the most asymmetrically arranged major capsid proteins yet observed in virus structures.
- Department of Biochemistry and Biophysics, Center for Phage Technology, Texas A&M University, College Station, TX 77843.
Organizational Affiliation: