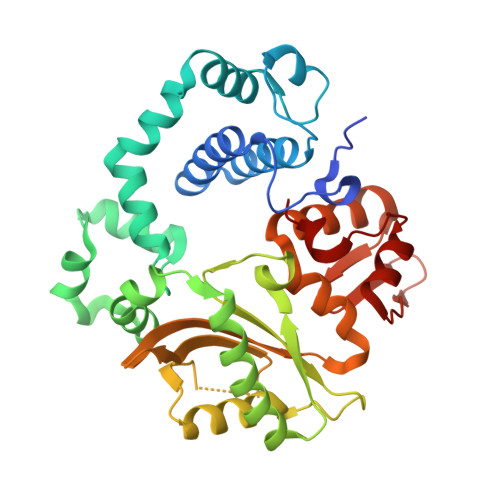
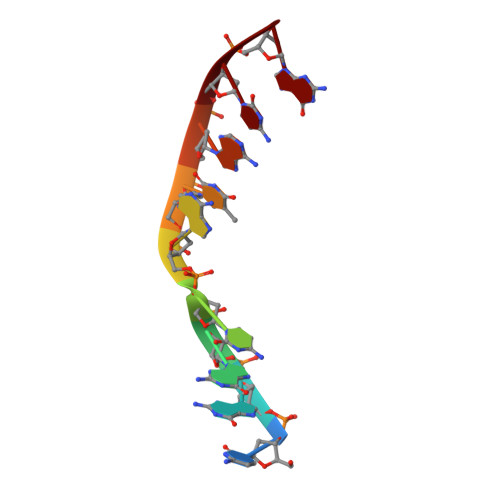
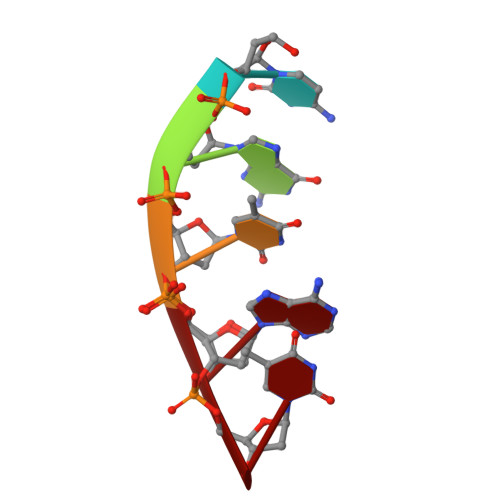
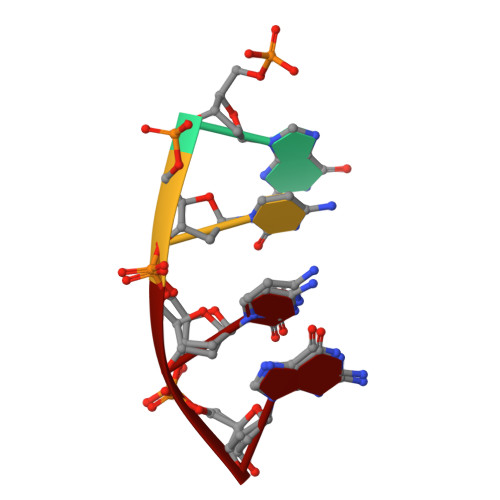
Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Jamsen, J.A., Beard, W.A., Pedersen, L.C., Shock, D.D., Moon, A.F., Krahn, J.M., Bebenek, K., Kunkel, T.A., Wilson, S.H.(2017) Nat Commun 8: 253-253
- PubMed: 28811466
- DOI: https://doi.org/10.1038/s41467-017-00271-7
- Primary Citation of Related Structures:
5TXX, 5TXZ, 5TYB, 5TYC, 5TYD, 5TYE, 5TYF, 5TYG, 5TYU, 5TYV, 5TYW, 5TYX, 5TYY, 5TYZ - PubMed Abstract:
DNA polymerase (pol) μ is a DNA-dependent polymerase that incorporates nucleotides during gap-filling synthesis in the non-homologous end-joining pathway of double-strand break repair. Here we report time-lapse X-ray crystallography snapshots of catalytic events during gap-filling DNA synthesis by pol μ. Unique catalytic intermediates and active site conformational changes that underlie catalysis are uncovered, and a transient third (product) metal ion is observed in the product state. The product manganese coordinates phosphate oxygens of the inserted nucleotide and PP i . The product metal is not observed during DNA synthesis in the presence of magnesium. Kinetic analyses indicate that manganese increases the rate constant for deoxynucleoside 5'-triphosphate insertion compared to magnesium. The likely product stabilization role of the manganese product metal in pol μ is discussed. These observations provide insight on structural attributes of this X-family double-strand break repair polymerase that impact its biological function in genome maintenance.DNA polymerase (pol) μ functions in DNA double-strand break repair. Here the authors use time-lapse X-ray crystallography to capture the states of pol µ during the conversion from pre-catalytic to product complex and observe a third transiently bound metal ion in the product state.
- Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC, 27709, USA.
Organizational Affiliation: