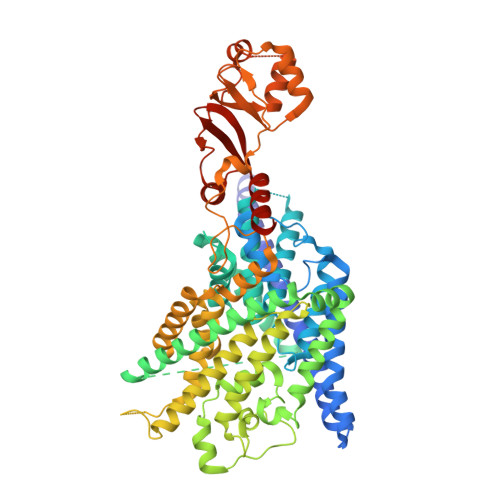
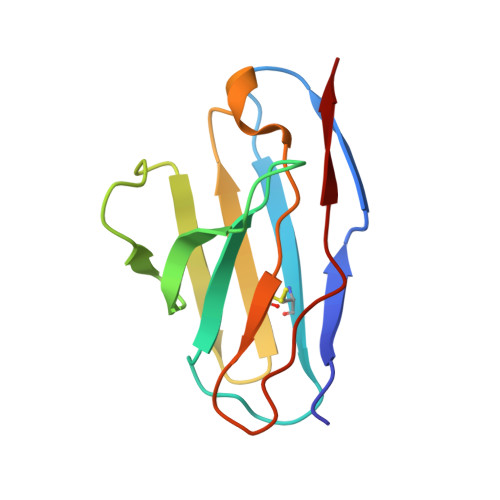
Structure of a CLC chloride ion channel by cryo-electron microscopy.
Park, E., Campbell, E.B., MacKinnon, R.(2017) Nature 541: 500-505
- PubMed: 28002411
- DOI: https://doi.org/10.1038/nature20812
- Primary Citation of Related Structures:
5TQQ, 5TR1 - PubMed Abstract:
CLC proteins transport chloride (Cl - ) ions across cellular membranes to regulate muscle excitability, electrolyte movement across epithelia, and acidification of intracellular organelles. Some CLC proteins are channels that conduct Cl - ions passively, whereas others are secondary active transporters that exchange two Cl - ions for one H + . The structural basis underlying these distinctive transport mechanisms is puzzling because CLC channels and transporters are expected to share the same architecture on the basis of sequence homology. Here we determined the structure of a bovine CLC channel (CLC-K) using cryo-electron microscopy. A conserved loop in the Cl - transport pathway shows a structure markedly different from that of CLC transporters. Consequently, the cytosolic constriction for Cl - passage is widened in CLC-K such that the kinetic barrier previously postulated for Cl - /H + transporter function would be reduced. Thus, reduction of a kinetic barrier in CLC channels enables fast flow of Cl - down its electrochemical gradient.
- Laboratory of Molecular Neurobiology and Biophysics and Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, New York 10065, USA.
Organizational Affiliation: