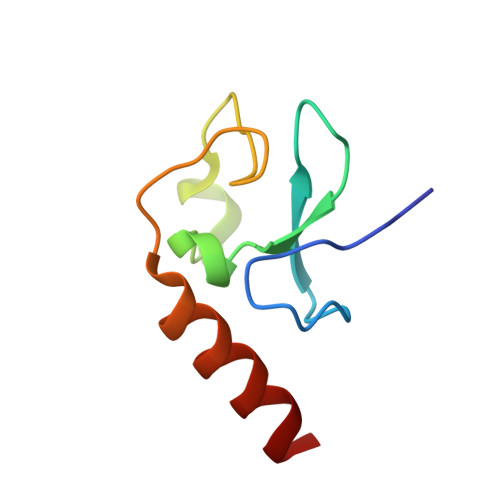
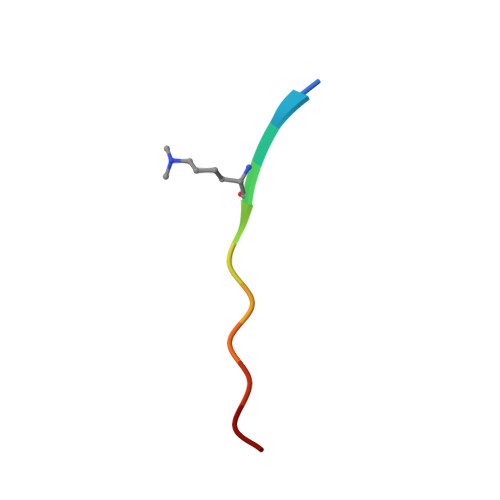
Structural Insight into Recognition of Methylated Histone H3K4 by Set3.
Gatchalian, J., Ali, M., Andrews, F.H., Zhang, Y., Barrett, A.S., Kutateladze, T.G.(2017) J Mol Biology 429: 2066-2074
- PubMed: 27697561
- DOI: https://doi.org/10.1016/j.jmb.2016.09.020
- Primary Citation of Related Structures:
5TDR, 5TDW - PubMed Abstract:
The plant homeodomain (PHD) finger of Set3 binds methylated lysine 4 of histone H3 in vitro and in vivo; however, precise selectivity of this domain has not been fully characterized. Here, we explore the determinants of methyllysine recognition by the PHD fingers of Set3 and its orthologs. We use X-ray crystallographic and spectroscopic approaches to show that the Set3 PHD finger binds di- and trimethylated states of H3K4 with comparable affinities and employs similar molecular mechanisms to form complexes with either mark. Composition of the methyllysine-binding pocket plays an essential role in determining the selectivity of the PHD fingers. The finding that the histone-binding activity is not conserved in the PHD finger of Set4 suggests different functions for the Set3 and Set4 paralogs.
- Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045, USA.
Organizational Affiliation: