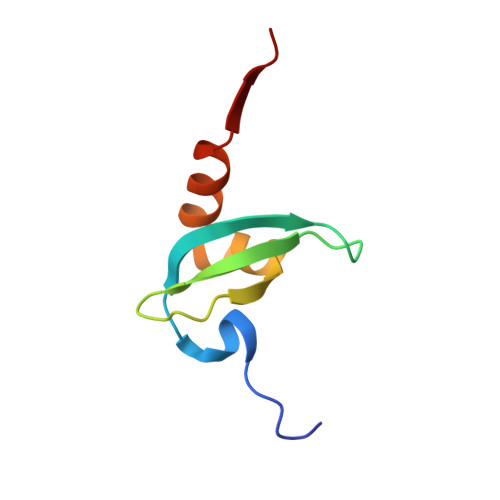
Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
Liu, Y., Qin, S., Lei, M., Tempel, W., Zhang, Y., Loppnau, P., Li, Y., Min, J.(2017) J Biological Chem 292: 5655-5664
- PubMed: 28223359
- DOI: https://doi.org/10.1074/jbc.M116.768374
- Primary Citation of Related Structures:
5T1I - PubMed Abstract:
Heterochromatin protein 1 (HP1), a highly conserved non-histone chromosomal protein in eukaryotes, plays important roles in the regulation of gene transcription. Each of the three human homologs of HP1 includes a chromoshadow domain (CSD). The CSD interacts with various proteins bearing the P X V X L motif but also with a region of histone H3 that bears the similar P XX V X L motif. The latter interaction has not yet been resolved in atomic detail. Here we demonstrate that the CSDs of all three human HP1 homologs have comparable affinities to the P XX V X L motif of histone H3. The HP1 C-terminal extension enhances the affinity, as does the increasing length of the H3 peptide. The crystal structure of the human HP1γ CSD (CSD γ ) in complex with an H3 peptide suggests that recognition of H3 by CSD γ to some extent resembles CSD-P X V X L interaction. Nevertheless, the prolyl residue of the P XX V X L motif appears to play a role distinct from that of Pro in the known HP1β CSD-P X V X L complexes. We consequently generalize the historical CSD-P X V X L interaction model and expand the search scope for additional CSD binding partners.
- From the Structural Genomics Consortium, University of Toronto, Toronto, Ontario M5G 1L7, Canada and.
Organizational Affiliation: