Structural and functional characterization of an electron transfer flavoprotein involved in toluene degradation in strictly anaerobic bacteria.
Vogt, M.S., Schuhle, K., Kolzer, S., Peschke, P., Chowdhury, N.P., Kleinsorge, D., Buckel, W., Essen, L.O., Heider, J.(2019) J Bacteriol
- PubMed: 31405915
- DOI: https://doi.org/10.1128/JB.00326-19
- Primary Citation of Related Structures:
5OW0 - PubMed Abstract:
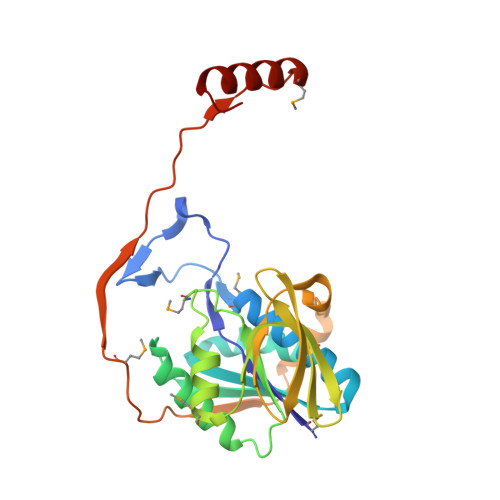
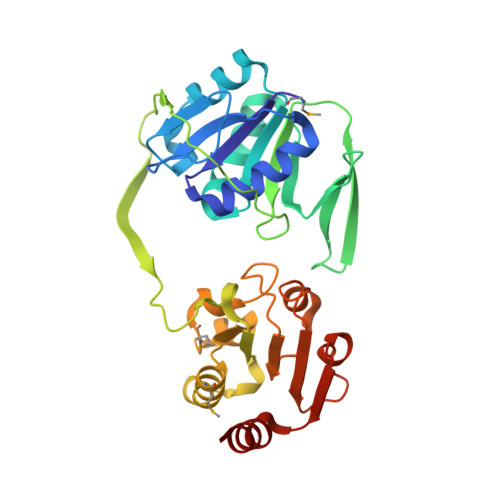
( R )-Benzylsuccinate is the characteristic initial intermediate of anaerobic toluene metabolism, which is formed by a radical-type addition of toluene to fumarate. Its further degradation proceeds by activation to the coenzyme A (CoA)-thioester and β-oxidation involving a specific ( R )-2-benzylsuccinyl-CoA dehydrogenase (BbsG) affiliated with the family of acyl-CoA dehydrogenases. In this report, we present the biochemical properties of electron transfer flavoproteins (ETFs) from the strictly anaerobic toluene-degrading species Geobacter metallireducens and Desulfobacula toluolica and the facultatively anaerobic bacterium Aromatoleum aromaticum We determined the X-ray structure of the ETF paralogue involved in toluene metabolism of G. metallireducens , revealing strong overall similarities to previously characterized ETF variants but significantly different structural properties in the hinge regions mediating conformational changes. We also show that all strictly anaerobic toluene degraders utilize one of multiple genome-encoded related ETF paralogues, which constitute a distinct clade of similar sequences in the ETF family, for β-oxidation of benzylsuccinate. In contrast, facultatively anaerobic toluene degraders contain only one ETF species, which is utilized in all β-oxidation pathways. Our phylogenetic analysis of the known sequences of the ETF family suggests that at least 36 different clades can be differentiated, which are defined either by the taxonomic group of the respective host species (e.g., clade P for Proteobacteria ) or by functional specialization (e.g., clade T for anaerobic toluene degradation). IMPORTANCE This study documents the involvement of ETF in anaerobic toluene metabolism as the physiological electron acceptor for benzylsuccinyl-CoA dehydrogenase. While toluene-degrading denitrifying proteobacteria use a common ETF species, which is also used for other β-oxidation pathways, obligately anaerobic sulfate- or ferric-iron-reducing bacteria use specialized ETF paralogues for toluene degradation. Based on the structure and sequence conservation of these ETFs, they form a new clade that is only remotely related to the previously characterized members of the ETF family. An exhaustive analysis of the available sequences indicated that the protein family consists of several closely related clades of proven or potential electron-bifurcating ETF species and many deeply branching nonbifurcating clades, which either follow the host phylogeny or are affiliated according to functional criteria.
- Faculty of Chemistry, Philipps-Universität Marburg, Marburg, Germany.
Organizational Affiliation: