Structure of a phosphoprotein-protein complex
Mukherjee, M., Bayliss, R.(2018) EMBO J
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Clathrin heavy chain 1 | 574 | Mus musculus | Mutation(s): 0 Gene Names: Cltc | 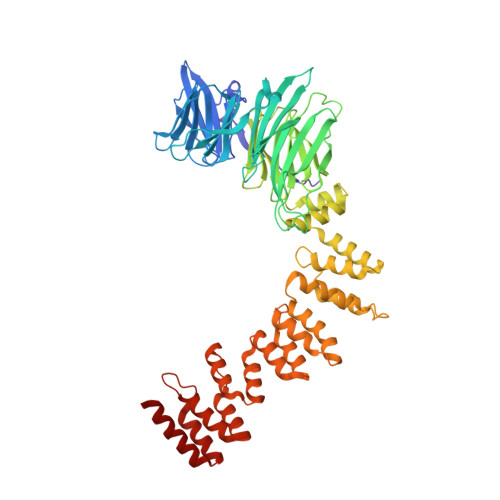 | |
UniProt | |||||
Find proteins for Q68FD5 (Mus musculus) Explore Q68FD5 Go to UniProtKB: Q68FD5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q68FD5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LYS-GLU-SER-ALA-LEU-ARG-LYS-GLN-SEP-LEU-TYR-LEU-LYS-PHE-ASP-PRO-LEU-LEU | 18 | Homo sapiens | Mutation(s): 0 | 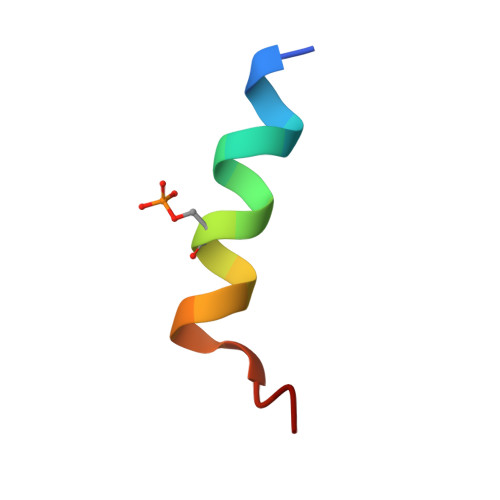 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y6A5 (Homo sapiens) Explore Q9Y6A5 Go to UniProtKB: Q9Y6A5 | |||||
PHAROS: Q9Y6A5 GTEx: ENSG00000013810 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y6A5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| SEP Query on SEP | E, F, G, H | L-PEPTIDE LINKING | C3 H8 N O6 P |  | SER |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 115.52 | α = 90 |
| b = 120.04 | β = 95.72 |
| c = 123.13 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| xia2 | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council | United Kingdom | BB/L023113/1 |