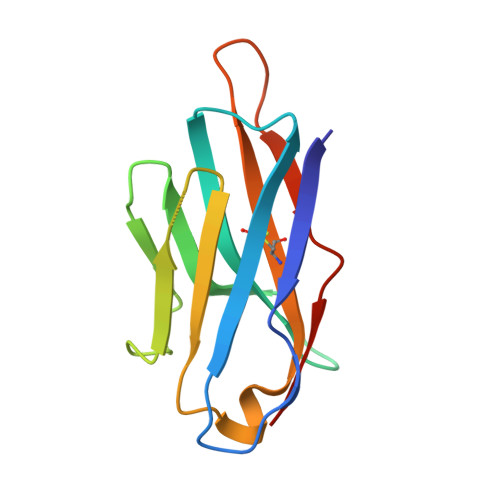
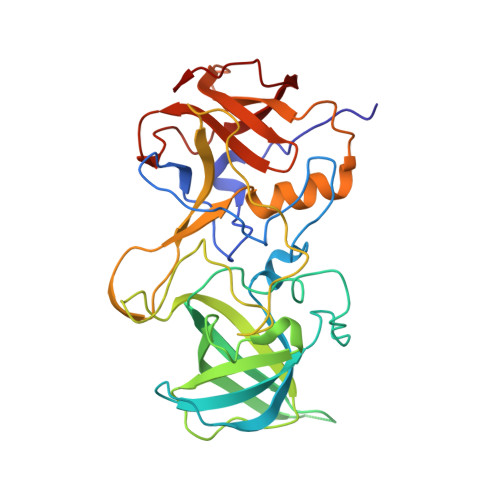
Nanobodies targeting norovirus capsid reveal functional epitopes and potential mechanisms of neutralization.
Koromyslova, A.D., Hansman, G.S.(2017) PLoS Pathog 13: e1006636-e1006636
- PubMed: 29095961
- DOI: https://doi.org/10.1371/journal.ppat.1006636
- Primary Citation of Related Structures:
5O02, 5O03, 5O04, 5O05, 5OMM, 5OMN - PubMed Abstract:
Norovirus is the leading cause of gastroenteritis worldwide. Despite recent developments in norovirus propagation in cell culture, these viruses are still challenging to grow routinely. Moreover, little is known on how norovirus infects the host cells, except that histo-blood group antigens (HBGAs) are important binding factors for infection and cell entry. Antibodies that bind at the HBGA pocket and block attachment to HBGAs are believed to neutralize the virus. However, additional neutralization epitopes elsewhere on the capsid likely exist and impeding the intrinsic structural dynamics of the capsid could be equally important. In the current study, we investigated a panel of Nanobodies in order to probe functional epitopes that could trigger capsid rearrangement and/ or interfere with HBGA binding interactions. The precise binding sites of six Nanobodies (Nano-4, Nano-14, Nano-26, Nano-27, Nano-32, and Nano-42) were identified using X-ray crystallography. We showed that these Nanobodies bound on the top, side, and bottom of the norovirus protruding domain. The impact of Nanobody binding on norovirus capsid morphology was analyzed using electron microscopy and dynamic light scattering. We discovered that distinct Nanobody epitopes were associated with varied changes in particle structural integrity and assembly. Interestingly, certain Nanobody-induced capsid morphological changes lead to the capsid protein degradation and viral RNA exposure. Moreover, Nanobodies employed multiple inhibition mechanisms to prevent norovirus attachment to HBGAs, which included steric obstruction (Nano-14), allosteric interference (Nano-32), and violation of normal capsid morphology (Nano-26 and Nano-85). Finally, we showed that two Nanobodies (Nano-26 and Nano-85) not only compromised capsid integrity and inhibited VLPs attachment to HBGAs, but also recognized a broad panel of norovirus genotypes with high affinities. Consequently, Nano-26 and Nano-85 have a great potential to function as novel therapeutic agents against human noroviruses.
- Schaller Research Group at the University of Heidelberg and the DKFZ, Heidelberg, Germany.
Organizational Affiliation: