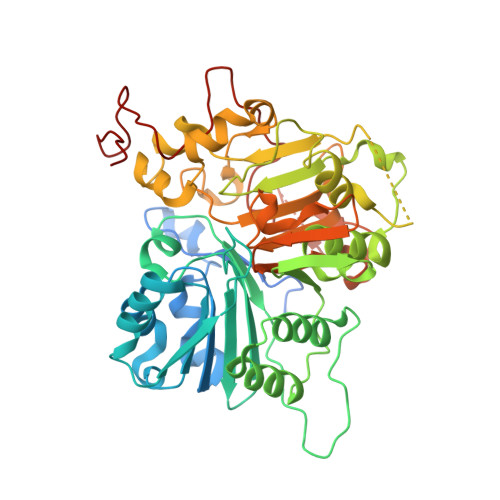
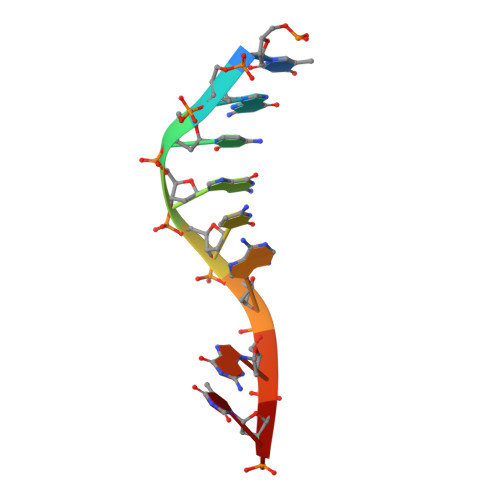
Structural basis for DNA 3'-end processing by human tyrosyl-DNA phosphodiesterase 1.
Flett, F.J., Ruksenaite, E., Armstrong, L.A., Bharati, S., Carloni, R., Morris, E.R., Mackay, C.L., Interthal, H., Richardson, J.M.(2018) Nat Commun 9: 24-24
- PubMed: 29295983
- DOI: https://doi.org/10.1038/s41467-017-02530-z
- Primary Citation of Related Structures:
5NW9, 5NWA - PubMed Abstract:
Tyrosyl-DNA phosphodiesterase (Tdp1) is a DNA 3'-end processing enzyme that repairs topoisomerase 1B-induced DNA damage. We use a new tool combining site-specific DNA-protein cross-linking with mass spectrometry to identify Tdp1 interactions with DNA. A conserved phenylalanine (F259) of Tdp1, required for efficient DNA processing in biochemical assays, cross-links to defined positions in DNA substrates. Crystal structures of Tdp1-DNA complexes capture the DNA repair machinery after 3'-end cleavage; these reveal how Tdp1 coordinates the 3'-phosphorylated product of nucleosidase activity and accommodates duplex DNA. A hydrophobic wedge splits the DNA ends, directing the scissile strand through a channel towards the active site. The F259 side-chain stacks against the -3 base pair, delimiting the junction of duplexed and melted DNA, and fixes the scissile strand in the channel. Our results explain why Tdp1 cleavage is non-processive and provide a molecular basis for DNA 3'-end processing by Tdp1.
- Institute of Cell Biology, School of Biological Sciences, University of Edinburgh, The King's Buildings, Roger Land Building, Alexander Crum Brown Road, Edinburgh, EH9 3FF, UK.
Organizational Affiliation: