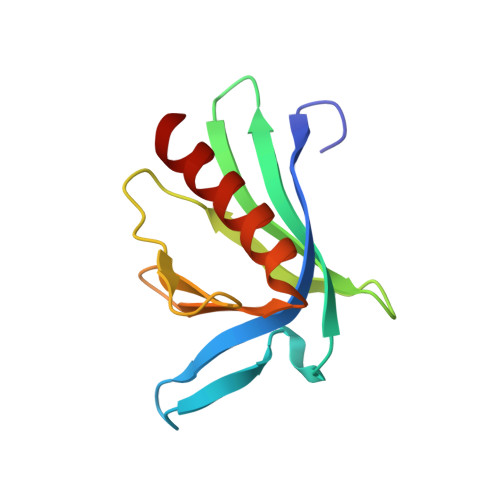
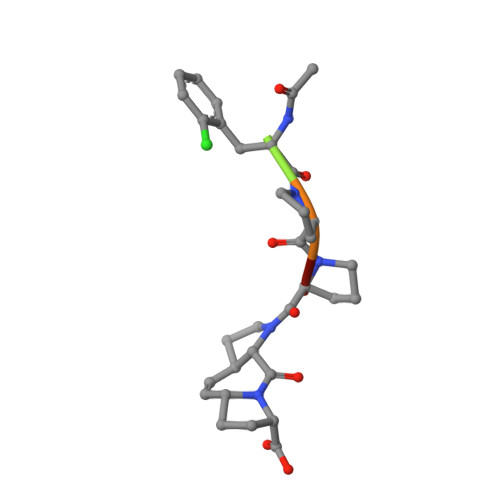
Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Barone, M., Muller, M., Chiha, S., Ren, J., Albat, D., Soicke, A., Dohmen, S., Klein, M., Bruns, J., van Dinther, M., Opitz, R., Lindemann, P., Beerbaum, M., Motzny, K., Roske, Y., Schmieder, P., Volkmer, R., Nazare, M., Heinemann, U., Oschkinat, H., Ten Dijke, P., Schmalz, H.G., Kuhne, R.(2020) Proc Natl Acad Sci U S A 117: 29684-29690
- PubMed: 33184177
- DOI: https://doi.org/10.1073/pnas.2007213117
- Primary Citation of Related Structures:
5N91, 5N9C, 5N9P, 5NAJ, 5NBF, 5NBX, 5NC2, 5NC7, 5NCF, 5NCG, 5NCP, 5ND0, 5NDU, 5NEG, 6RCF, 6RCJ, 6RD2, 6XVT, 6XXR, 7A5M, 7AKI - PubMed Abstract:
Battling metastasis through inhibition of cell motility is considered a promising approach to support cancer therapies. In this context, Ena/VASP-depending signaling pathways, in particular interactions with their EVH1 domains, are promising targets for pharmaceutical intervention. However, protein-protein interactions involving proline-rich segments are notoriously difficult to address by small molecules. Hence, structure-based design efforts in combination with the chemical synthesis of additional molecular entities are required. Building on a previously developed nonpeptidic micromolar inhibitor, we determined 22 crystal structures of ENAH EVH1 in complex with inhibitors and rationally extended our library of conformationally defined proline-derived modules (ProMs) to succeed in developing a nanomolar inhibitor ([Formula: see text] Da). In contrast to the previous inhibitor, the optimized compounds reduced extravasation of invasive breast cancer cells in a zebrafish model. This study represents an example of successful, structure-guided development of low molecular weight inhibitors specifically and selectively addressing a proline-rich sequence-recognizing domain that is characterized by a shallow epitope lacking defined binding pockets. The evolved high-affinity inhibitor may now serve as a tool in validating the basic therapeutic concept, i.e., the suppression of cancer metastasis by inhibiting a crucial protein-protein interaction involved in actin filament processing and cell migration.
- Department of Structural Biology, Leibniz-Forschungsinstitut für Molekulare Pharmakologie, 13125 Berlin, Germany.
Organizational Affiliation: