Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Chen, W.F., Rety, S., Guo, H.L., Dai, Y.X., Wu, W.Q., Liu, N.N., Auguin, D., Liu, Q.W., Hou, X.M., Dou, S.X., Xi, X.G.(2018) Structure 26: 403-415.e4
- PubMed: 29429875
- DOI: https://doi.org/10.1016/j.str.2018.01.008
- Primary Citation of Related Structures:
5N8R, 5N8S, 5N90, 5N94, 5N96, 5N98, 5N9A, 5N9D - PubMed Abstract:
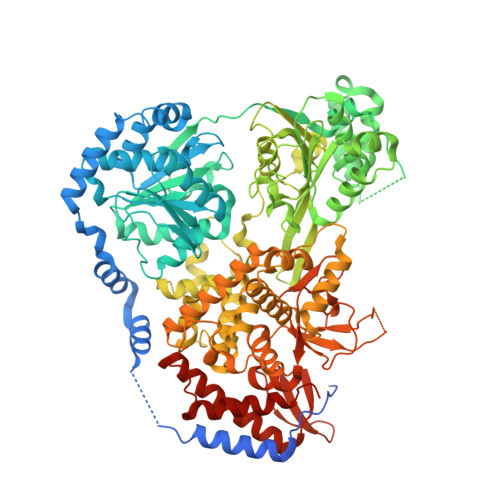
Helicase DHX36 plays essential roles in cell development and differentiation at least partially by resolving G-quadruplex (G4) structures. Here we report crystal structures of the Drosophila homolog of DHX36 (DmDHX36) in complex with RNA and a series of DNAs. By combining structural, small-angle X-ray scattering, molecular dynamics simulation, and single-molecule fluorescence studies, we revealed that positively charged amino acids in RecA2 and OB-like domains constitute an elaborate structural pocket at the nucleic acid entrance, in which negatively charged G4 DNA is tightly bound and partially destabilized. The G4 DNA is then completely unfolded through the 3'-5' translocation activity of the helicase. Furthermore, crystal structures and DNA binding assays show that G-rich DNA is preferentially recognized and in the presence of ATP, specifically bound by DmDHX36, which may cooperatively enhance the G-rich DNA translocation and G4 unfolding. On the basis of these results, a conceptual G4 DNA-resolving mechanism is proposed.
- College of Life Sciences, Northwest A&F University, Yangling, Shaanxi 712100, China.
Organizational Affiliation: