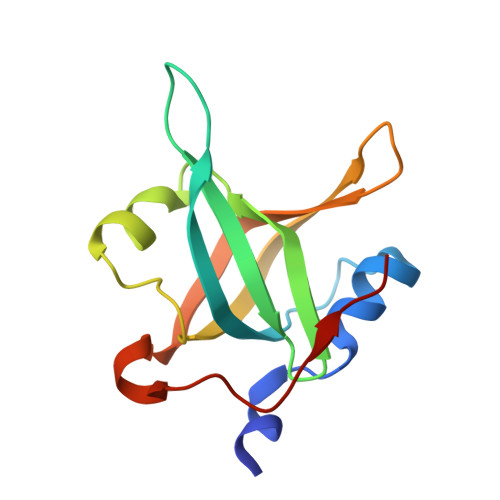
Molecular basis for PrimPol recruitment to replication forks by RPA.
Guilliam, T.A., Brissett, N.C., Ehlinger, A., Keen, B.A., Kolesar, P., Taylor, E.M., Bailey, L.J., Lindsay, H.D., Chazin, W.J., Doherty, A.J.(2017) Nat Commun 8: 15222-15222
- PubMed: 28534480
- DOI: https://doi.org/10.1038/ncomms15222
- Primary Citation of Related Structures:
5N85, 5N8A - PubMed Abstract:
DNA damage and secondary structures can stall the replication machinery. Cells possess numerous tolerance mechanisms to complete genome duplication in the presence of such impediments. In addition to translesion synthesis (TLS) polymerases, most eukaryotic cells contain a multifunctional replicative enzyme called primase-polymerase (PrimPol) that is capable of directly bypassing DNA damage by TLS, as well as repriming replication downstream of impediments. Here, we report that PrimPol is recruited to reprime through its interaction with RPA. Using biophysical and crystallographic approaches, we identify that PrimPol possesses two RPA-binding motifs and ascertained the key residues required for these interactions. We demonstrate that one of these motifs is critical for PrimPol's recruitment to stalled replication forks in vivo. In addition, biochemical analysis reveals that RPA serves to stimulate the primase activity of PrimPol. Together, these findings provide significant molecular insights into PrimPol's mode of recruitment to stalled forks to facilitate repriming and restart.
- Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Brighton BN1 9RQ, UK.
Organizational Affiliation: