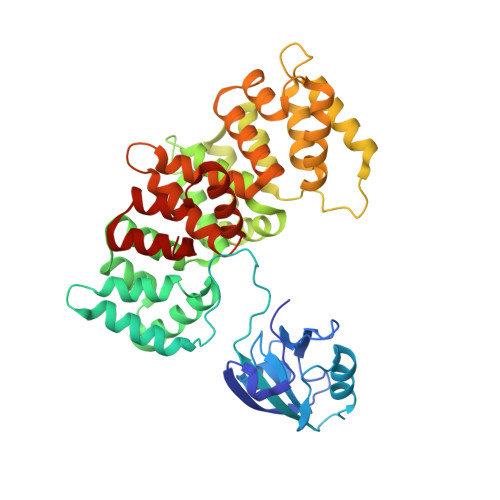
Dynamic control of RSK complexes by phosphoswitch-based regulation.
Gogl, G., Biri-Kovacs, B., Poti, A.L., Vadaszi, H., Szeder, B., Bodor, A., Schlosser, G., Acs, A., Turiak, L., Buday, L., Alexa, A., Nyitray, L., Remenyi, A.(2018) FEBS J 285: 46-71
- PubMed: 29083550
- DOI: https://doi.org/10.1111/febs.14311
- Primary Citation of Related Structures:
5N7D, 5N7F, 5N7G - PubMed Abstract:
Assembly and disassembly of protein-protein complexes needs to be dynamically controlled and phosphoswitches based on linear motifs are crucial in this process. Extracellular signal-regulated kinase 2 (ERK2) recognizes a linear-binding motif at the C-terminal tail (CTT) of ribosomal S6 kinase 1 (RSK1), leading to phosphorylation and subsequent activation of RSK1. The CTT also contains a classical PDZ domain-binding motif which binds RSK substrates (e.g. MAGI-1). We show that autophosphorylation of the disordered CTT promotes the formation of an intramolecular charge clamp, which efficiently masks critical residues and indirectly hinders ERK binding. Thus, RSK1 CTT operates as an autoregulated phosphoswitch: its phosphorylation at specific sites affects its protein-binding capacity and its conformational dynamics. These biochemical feedbacks, which form the structural basis for the rapid dissociation of ERK2-RSK1 and RSK1-PDZ substrate complexes under sustained epidermal growth factor (EGF) stimulation, were structurally characterized and validated in living cells. Overall, conformational changes induced by phosphorylation in disordered regions of protein kinases, coupled to allosteric events occurring in the kinase domain cores, may provide mechanisms that contribute to the emergence of complex signaling activities. In addition, we show that phosphoswitches based on linear motifs can be functionally classified as ON and OFF protein-protein interaction switches or dimmers, depending on the specific positioning of phosphorylation target sites in relation to functional linear-binding motifs. Moreover, interaction of phosphorylated residues with positively charged residues in disordered regions is likely to be a common mechanism of phosphoregulation. Structural data are available in the PDB database under the accession numbers 5N7D, 5N7F and 5N7G. NMR spectral assignation data are available in the BMRB database under the accession numbers 27213 and 27214.
- Department of Biochemistry, ELTE Eötvös Loránd University, Budapest, Hungary.
Organizational Affiliation: