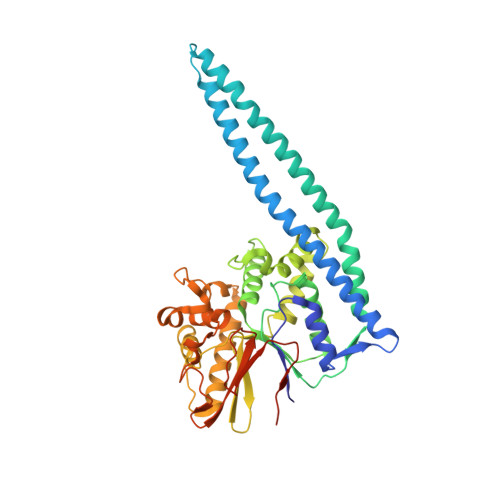
Structure of the WipA protein reveals a novel tyrosine protein phosphatase effector from Legionella pneumophila.
Pinotsis, N., Waksman, G.(2017) J Biological Chem 292: 9240-9251
- PubMed: 28389563
- DOI: https://doi.org/10.1074/jbc.M117.781948
- Primary Citation of Related Structures:
5N6X, 5N72 - PubMed Abstract:
Legionnaires' disease is a severe form of pneumonia caused by the bacterium Legionella pneumophila. L. pneumophila pathogenicity relies on secretion of more than 300 effector proteins by a type IVb secretion system. Among these Legionella effectors, WipA has been primarily studied because of its dependence on a chaperone complex, IcmSW, for translocation through the secretion system, but its role in pathogenicity has remained unknown. In this study, we present the crystal structure of a large fragment of WipA, WipA435. Surprisingly, this structure revealed a serine/threonine phosphatase fold that unexpectedly targets tyrosine-phosphorylated peptides. The structure also revealed a sequence insertion that folds into an α-helical hairpin, the tip of which adopts a canonical coiled-coil structure. The purified protein was a dimer whose dimer interface involves interactions between the coiled coil of one WipA molecule and the phosphatase domain of another. Given the ubiquity of protein-protein interaction mediated by interactions between coiled-coils, we hypothesize that WipA can thereby transition from a homodimeric state to a heterodimeric state in which the coiled-coil region of WipA is engaged in a protein-protein interaction with a tyrosine-phosphorylated host target. In conclusion, these findings help advance our understanding of the molecular mechanisms of an effector involved in Legionella virulence and may inform approaches to elucidate the function of other effectors.
- From the Department of Biological Sciences, Institute of Structural and Molecular Biology, Birkbeck, Malet Street, WC1E 7HX London, United Kingdom and.
Organizational Affiliation: