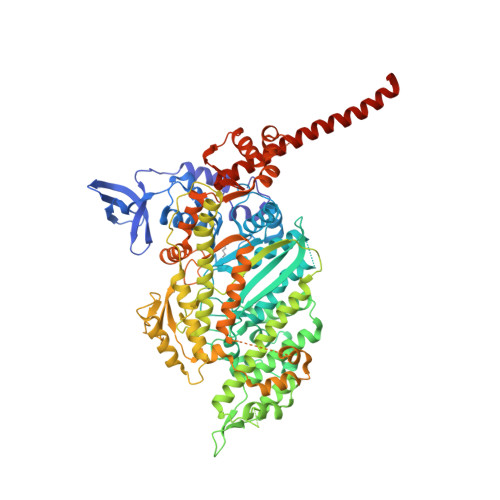
Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil.
Planelles-Herrero, V.J., Hartman, J.J., Robert-Paganin, J., Malik, F.I., Houdusse, A.(2017) Nat Commun 8: 190-190
- PubMed: 28775348
- DOI: https://doi.org/10.1038/s41467-017-00176-5
- Primary Citation of Related Structures:
5N69, 5N6A - PubMed Abstract:
Omecamtiv mecarbil is a selective, small-molecule activator of cardiac myosin that is being developed as a potential treatment for heart failure with reduced ejection fraction. Here we determine the crystal structure of cardiac myosin in the pre-powerstroke state, the most relevant state suggested by kinetic studies, both with (2.45 Å) and without (3.10 Å) omecamtiv mecarbil bound. Omecamtiv mecarbil does not change the motor mechanism nor does it influence myosin structure. Instead, omecamtiv mecarbil binds to an allosteric site that stabilizes the lever arm in a primed position resulting in accumulation of cardiac myosin in the primed state prior to onset of cardiac contraction, thus increasing the number of heads that can bind to the actin filament and undergo a powerstroke once the cardiac cycle starts. The mechanism of action of omecamtiv mecarbil also provides insights into uncovering how force is generated by molecular motors.Omecamtiv mecarbil (OM) is a cardiac myosin activator that is currently in clinical trials for heart failure treatment. Here, the authors give insights into its mode of action and present the crystal structure of OM bound to bovine cardiac myosin, which shows that OM stabilizes the pre-powerstroke state of myosin.
- Structural Motility, Institut Curie, PSL Research University, CNRS, UMR 144, F-75005, Paris, France.
Organizational Affiliation: