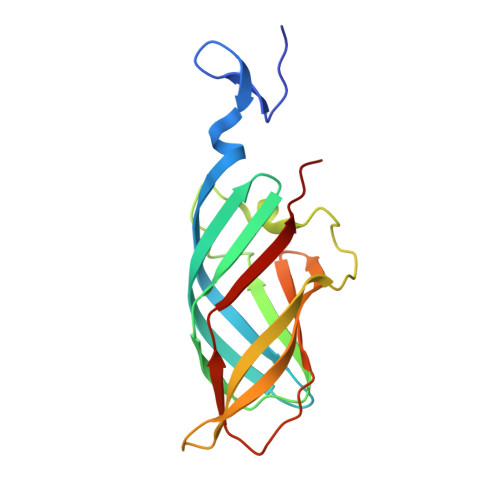
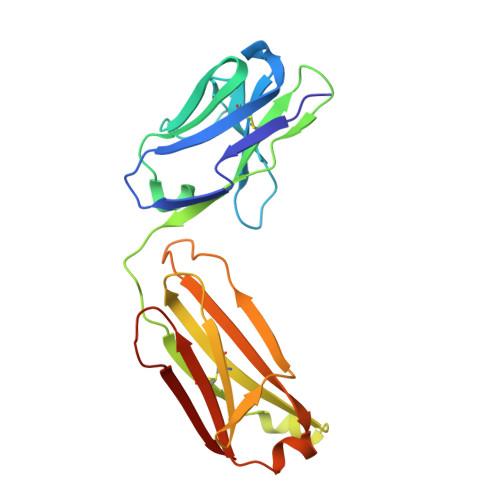
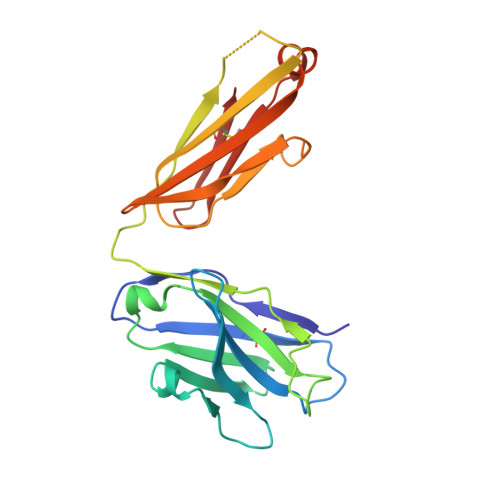
Structural Insights into Reovirus sigma 1 Interactions with Two Neutralizing Antibodies.
Dietrich, M.H., Ogden, K.M., Katen, S.P., Reiss, K., Sutherland, D.M., Carnahan, R.H., Goff, M., Cooper, T., Dermody, T.S., Stehle, T.(2017) J Virol 91
- PubMed: 27928010
- DOI: https://doi.org/10.1128/JVI.01621-16
- Primary Citation of Related Structures:
5MHR, 5MHS - PubMed Abstract:
Reovirus attachment protein σ1 engages glycan receptors and junctional adhesion molecule-A (JAM-A) and is thought to undergo a conformational change during the proteolytic disassembly of virions to infectious subvirion particles (ISVPs) that accompanies cell entry. The σ1 protein is also the primary target of neutralizing antibodies. Here, we present a structural and functional characterization of two neutralizing antibodies that target σ1 of serotype 1 (T1) and serotype 3 (T3) reoviruses. The crystal structures revealed that each antibody engages its cognate σ1 protein within the head domain via epitopes distinct from the JAM-A-binding site. Surface plasmon resonance and cell-binding assays indicated that both antibodies likely interfere with JAM-A engagement by steric hindrance. To define the interplay between the carbohydrate receptor and antibody binding, we conducted hemagglutination inhibition assays using virions and ISVPs. The glycan-binding site of T1 σ1 is located in the head domain and is partly occluded by the bound Fab in the crystal structure. The T1-specific antibody inhibited hemagglutination by virions and ISVPs, probably via direct interference with glycan engagement. In contrast to T1 σ1, the carbohydrate-binding site of T3 σ1 is located in the tail domain, distal to the antibody epitope. The T3-specific antibody inhibited hemagglutination by T3 virions but not ISVPs, indicating that the antibody- and glycan-binding sites in σ1 are in closer spatial proximity on virions than on ISVPs. Our results provide direct evidence for a structural rearrangement of σ1 during virion-to-ISVP conversion and contribute new information about the mechanisms of antibody-mediated neutralization of reovirus. Virus attachment proteins mediate binding to host cell receptors, serve critical functions in cell and tissue tropism, and are often targeted by the neutralizing antibody response. The structural investigation of antibody-antigen complexes can provide valuable information for understanding the molecular basis of virus neutralization. Studies with enveloped viruses, such as HIV and influenza virus, have helped to define sites of vulnerability and guide vaccination strategies. By comparison, less is known about antibody binding to nonenveloped viruses. Here, we structurally investigated two neutralizing antibodies that bind the attachment protein σ1 of reovirus. Furthermore, we characterized the neutralization efficiency, the binding affinity for σ1, and the effect of the antibodies on reovirus receptor engagement. Our analysis defines reovirus interactions with two neutralizing antibodies, allows us to propose a mechanism by which they block virus infection, and provides evidence for a conformational change in the σ1 protein during viral cell entry.
- Interfaculty Institute of Biochemistry, University of Tübingen, Tübingen, Germany.
Organizational Affiliation: