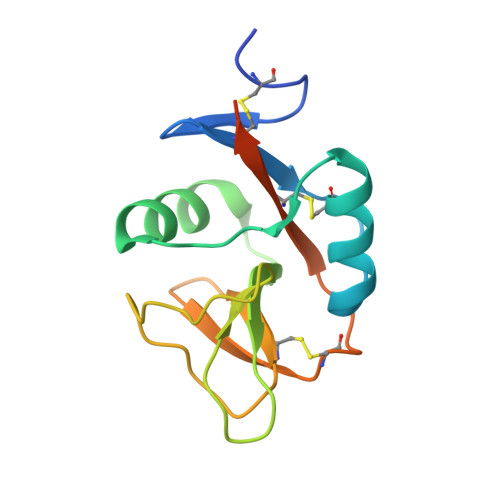
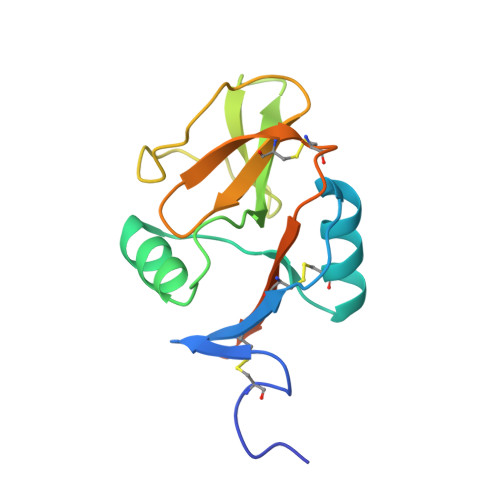
Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Blaha, J., Skalova, T., Kalouskova, B., Skorepa, O., Cmunt, D., Grobarova, V., Pazicky, S., Polachova, E., Abreu, C., Stransky, J., Koval, T., Duskova, J., Zhao, Y., Harlos, K., Hasek, J., Dohnalek, J., Vanek, O.(2022) Nat Commun 13: 5022-5022
- PubMed: 36028489
- DOI: https://doi.org/10.1038/s41467-022-32577-6
- Primary Citation of Related Structures:
5MGR, 5MGS, 5MGT - PubMed Abstract:
Signaling by the human C-type lectin-like receptor, natural killer (NK) cell inhibitory receptor NKR-P1, has a critical role in many immune-related diseases and cancer. C-type lectin-like receptors have weak affinities to their ligands; therefore, setting up a comprehensive model of NKR-P1-LLT1 interactions that considers the natural state of the receptor on the cell surface is necessary to understand its functions. Here we report the crystal structures of the NKR-P1 and NKR-P1:LLT1 complexes, which provides evidence that NKR-P1 forms homodimers in an unexpected arrangement to enable LLT1 binding in two modes, bridging two LLT1 molecules. These interaction clusters are suggestive of an inhibitory immune synapse. By observing the formation of these clusters in solution using SEC-SAXS analysis, by dSTORM super-resolution microscopy on the cell surface, and by following their role in receptor signaling with freshly isolated NK cells, we show that only the ligation of both LLT1 binding interfaces leads to effective NKR-P1 inhibitory signaling. In summary, our findings collectively support a model of NKR-P1:LLT1 clustering, which allows the interacting proteins to overcome weak ligand-receptor affinity and to trigger signal transduction upon cellular contact in the immune synapse.
- Department of Biochemistry, Faculty of Science, Charles University, Hlavova 2030, 12800, Prague, Czech Republic.
Organizational Affiliation: