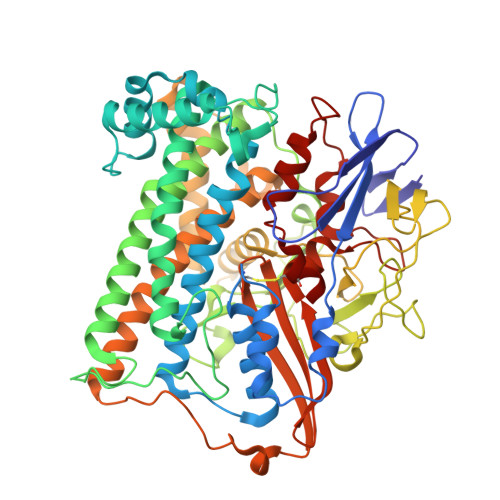
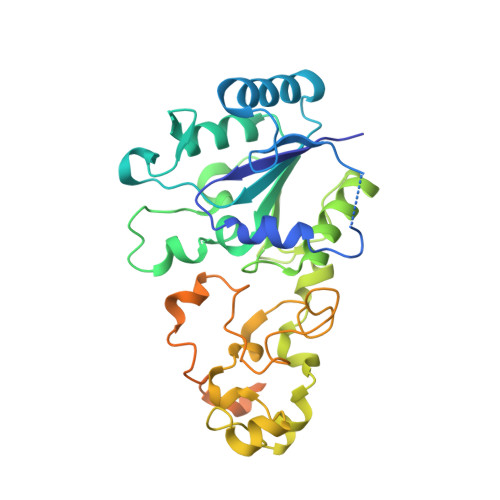
Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Kalms, J., Schmidt, A., Frielingsdorf, S., Utesch, T., Gotthard, G., von Stetten, D., van der Linden, P., Royant, A., Mroginski, M.A., Carpentier, P., Lenz, O., Scheerer, P.(2018) Proc Natl Acad Sci U S A 115: E2229-E2237
- PubMed: 29463722
- DOI: https://doi.org/10.1073/pnas.1712267115
- Primary Citation of Related Structures:
5MDJ, 5MDK, 5MDL - PubMed Abstract:
[NiFe] hydrogenases catalyze the reversible splitting of H 2 into protons and electrons at a deeply buried active site. The catalytic center can be accessed by gas molecules through a hydrophobic tunnel network. While most [NiFe] hydrogenases are inactivated by O 2 , a small subgroup, including the membrane-bound [NiFe] hydrogenase (MBH) of Ralstonia eutropha , is able to overcome aerobic inactivation by catalytic reduction of O 2 to water. This O 2 tolerance relies on a special [4Fe3S] cluster that is capable of releasing two electrons upon O 2 attack. Here, the O 2 accessibility of the MBH gas tunnel network has been probed experimentally using a "soak-and-freeze" derivatization method, accompanied by protein X-ray crystallography and computational studies. This combined approach revealed several sites of O 2 molecules within a hydrophobic tunnel network leading, via two tunnel entrances, to the catalytic center of MBH. The corresponding site occupancies were related to the O 2 concentrations used for MBH crystal derivatization. The examination of the O 2 -derivatized data furthermore uncovered two unexpected structural alterations at the [4Fe3S] cluster, which might be related to the O 2 tolerance of the enzyme.
- Charité - Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Institute of Medical Physics and Biophysics, Group Protein X-ray Crystallography and Signal Transduction, D-10117 Berlin, Germany.
Organizational Affiliation: