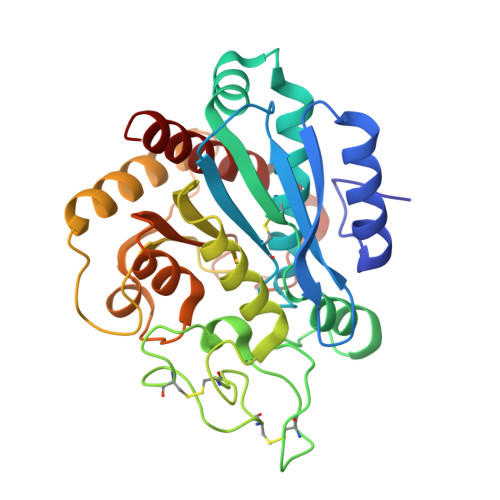
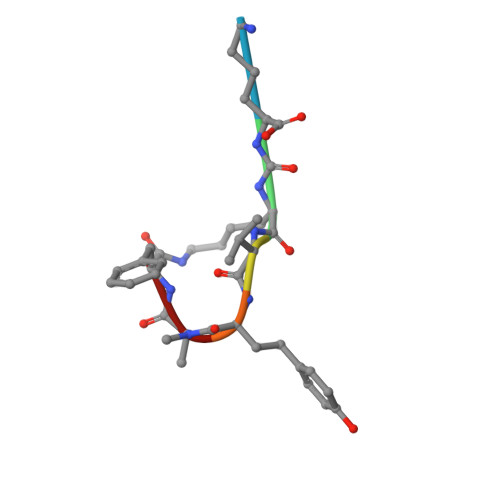
Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Schreuder, H., Liesum, A., Lonze, P., Stump, H., Hoffmann, H., Schiell, M., Kurz, M., Toti, L., Bauer, A., Kallus, C., Klemke-Jahn, C., Czech, J., Kramer, D., Enke, H., Niedermeyer, T.H., Morrison, V., Kumar, V., Bronstrup, M.(2016) Sci Rep 6: 32958-32958
- PubMed: 27604544
- DOI: https://doi.org/10.1038/srep32958
- Primary Citation of Related Structures:
5LRG, 5LRJ, 5LRK - PubMed Abstract:
Mature thrombin activatable fibrinolysis inhibitor (TAFIa) is a carboxypeptidase that stabilizes fibrin clots by removing C-terminal arginines and lysines from partially degraded fibrin. Inhibition of TAFIa stimulates the degradation of fibrin clots and may help to prevent thrombosis. Applying a lead finding approach based on literature-mining, we discovered that anabaenopeptins, cyclic peptides produced by cyanobacteria, were potent inhibitors of TAFIa with IC50 values as low as 1.5 nM. We describe the isolation and structure elucidation of 20 anabaenopeptins, including 13 novel congeners, as well as their pronounced structure-activity relationships (SAR) with respect to inhibition of TAFIa. Crystal structures of the anabaenopeptins B, C and F bound to the surrogate protease carboxypeptidase B revealed the binding modes of these large (~850 Da) compounds in detail and explained the observed SAR, i.e. the strong dependence of the potency on a basic (Arg, Lys) exocyclic residue that addressed the S1' binding pocket, and a broad tolerance towards substitutions in the pentacyclic ring that acted as a plug of the active site.
- Sanofi-Aventis Deutschland GmbH, Industriepark Höchst, 65926, Frankfurt am Main, Germany.
Organizational Affiliation: