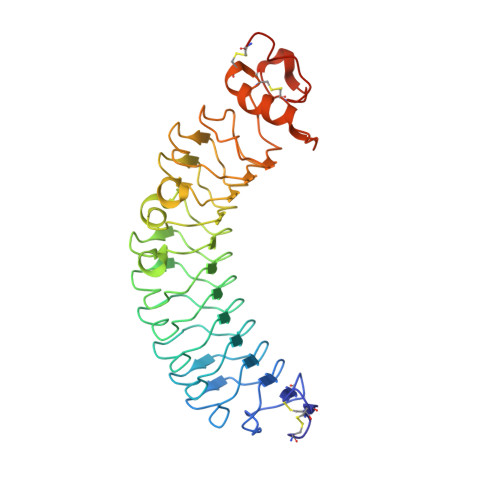
Crystal structure of human chondroadherin: solving a difficult molecular-replacement problem using de novo models.
Ramisch, S., Pramhed, A., Tillgren, V., Aspberg, A., Logan, D.T.(2017) Acta Crystallogr D Struct Biol 73: 53-63
- PubMed: 28045385
- DOI: https://doi.org/10.1107/S205979831601980X
- Primary Citation of Related Structures:
5LFN - PubMed Abstract:
Chondroadherin (CHAD) is a cartilage matrix protein that mediates the adhesion of isolated chondrocytes. Its protein core is composed of 11 leucine-rich repeats (LRR) flanked by cysteine-rich domains. CHAD makes important interactions with collagen as well as with cell-surface heparin sulfate proteoglycans and α 2 β 1 integrins. The integrin-binding site is located in a region of hitherto unknown structure at the C-terminal end of CHAD. Peptides based on the C-terminal human CHAD (hCHAD) sequence have shown therapeutic potential for treating osteoporosis. This article describes a still-unconventional structure solution by phasing with de novo models, the first of a β-rich protein. Structure determination of hCHAD using traditional, though nonsystematic, molecular replacement was unsuccessful in the hands of the authors, possibly owing to a combination of low sequence identity to other LRR proteins, four copies in the asymmetric unit and weak translational pseudosymmetry. However, it was possible to solve the structure by generating a large number of de novo models for the central LRR domain using Rosetta and multiple parallel molecular-replacement attempts using AMPLE. The hCHAD structure reveals an ordered C-terminal domain belonging to the LRRCT fold, with the integrin-binding motif (WLEAK) being part of a regular α-helix, and suggests ways in which experimental therapeutic peptides can be improved. The crystal structure itself and docking simulations further support that hCHAD dimers form in a similar manner to other matrix LRR proteins.
- Department of Biochemistry and Structural Biology, Lund University, Box 124, SE-221 00 Lund, Sweden.
Organizational Affiliation: