Structure and Stability of FlgD from the Pathogenic 26695 Strain of Helicobacter pylori
Kekez, I., Cendron, L., Stojanovic, M., Zanotti, G., Matkovic-Calogovic, D.(2016) Croat Chem Acta
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2016) Croat Chem Acta
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Basal-body rod modification protein FlgD | 146 | Helicobacter pylori 26695 | Mutation(s): 0 Gene Names: HP_0907 | 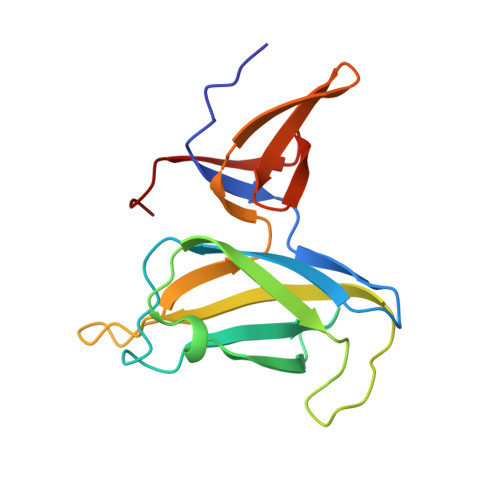 | |
UniProt | |||||
Find proteins for O25565 (Helicobacter pylori (strain ATCC 700392 / 26695)) Explore O25565 Go to UniProtKB: O25565 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O25565 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 77.51 | α = 90 |
| b = 34 | β = 98.9 |
| c = 133.34 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| iMOSFLM | data reduction |
| SCALA | data scaling |
| AMoRE | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| University of Padua | Italy | PRIN 2010-2011 (MIUR) |