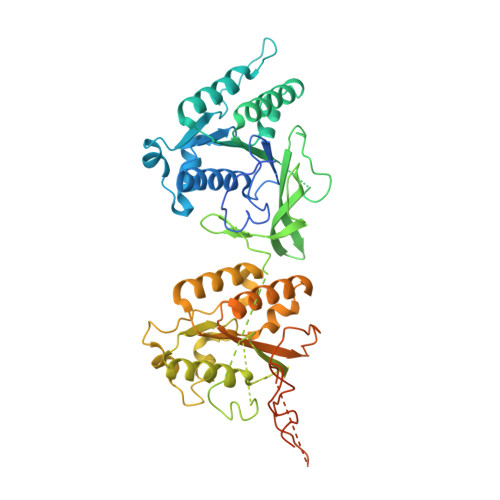
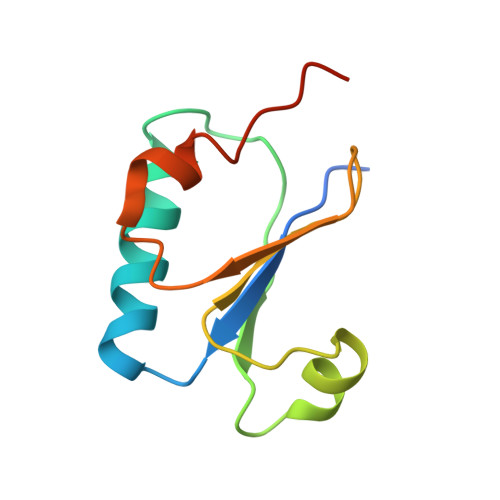
Structural basis of the day-night transition in a bacterial circadian clock.
Tseng, R., Goularte, N.F., Chavan, A., Luu, J., Cohen, S.E., Chang, Y.G., Heisler, J., Li, S., Michael, A.K., Tripathi, S., Golden, S.S., LiWang, A., Partch, C.L.(2017) Science 355: 1174-1180
- PubMed: 28302851
- DOI: https://doi.org/10.1126/science.aag2516
- Primary Citation of Related Structures:
5JWO, 5JWQ, 5JWR, 5JYT, 5JYU, 5JYV - PubMed Abstract:
Circadian clocks are ubiquitous timing systems that induce rhythms of biological activities in synchrony with night and day. In cyanobacteria, timing is generated by a posttranslational clock consisting of KaiA, KaiB, and KaiC proteins and a set of output signaling proteins, SasA and CikA, which transduce this rhythm to control gene expression. Here, we describe crystal and nuclear magnetic resonance structures of KaiB-KaiC,KaiA-KaiB-KaiC, and CikA-KaiB complexes. They reveal how the metamorphic properties of KaiB, a protein that adopts two distinct folds, and the post-adenosine triphosphate hydrolysis state of KaiC create a hub around which nighttime signaling events revolve, including inactivation of KaiA and reciprocal regulation of the mutually antagonistic signaling proteins, SasA and CikA.
- Quantitative and Systems Biology, University of California, Merced, CA 95343, USA.
Organizational Affiliation: