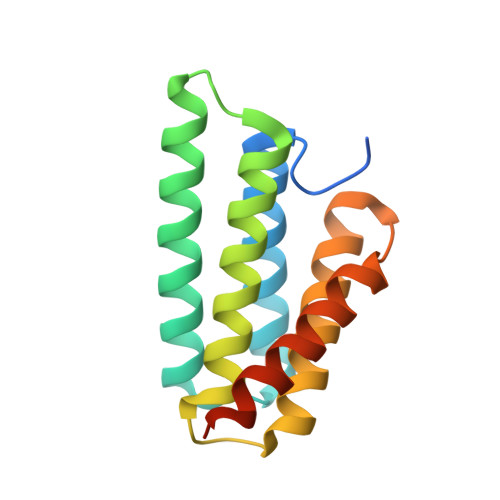
Crystal Structure of the Nephila clavipes Major Ampullate Spidroin 1A N-terminal Domain Reveals Plasticity at the Dimer Interface.
Atkison, J.H., Parnham, S., Marcotte, W.R., Olsen, S.K.(2016) J Biological Chem 291: 19006-19017
- PubMed: 27445329
- DOI: https://doi.org/10.1074/jbc.M116.736710
- Primary Citation of Related Structures:
5IZ2 - PubMed Abstract:
Spider dragline silk is a natural polymer harboring unique physical and biochemical properties that make it an ideal biomaterial. Artificial silk production requires an understanding of the in vivo mechanisms spiders use to convert soluble proteins, called spidroins, into insoluble fibers. Controlled dimerization of the spidroin N-terminal domain (NTD) is crucial to this process. Here, we report the crystal structure of the Nephila clavipes major ampullate spidroin NTD dimer. Comparison of our N. clavipes NTD structure with previously determined Euprosthenops australis NTD structures reveals subtle conformational alterations that lead to differences in how the subunits are arranged at the dimer interface. We observe a subset of contacts that are specific to each ortholog, as well as a substantial increase in asymmetry in the interactions observed at the N. clavipes NTD dimer interface. These asymmetric interactions include novel intermolecular salt bridges that provide new insights into the mechanism of NTD dimerization. We also observe a unique intramolecular "handshake" interaction between two conserved acidic residues that our data suggest adds an additional layer of complexity to the pH-sensitive relay mechanism for NTD dimerization. The results of a panel of tryptophan fluorescence dimerization assays probing the importance of these interactions support our structural observations. Based on our findings, we propose that conformational selectivity and plasticity at the NTD dimer interface play a role in the pH-dependent transition of the NTD from monomer to stably associated dimer as the spidroin progresses through the silk extrusion duct.
- From the Department of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, South Carolina 29425 and.
Organizational Affiliation: