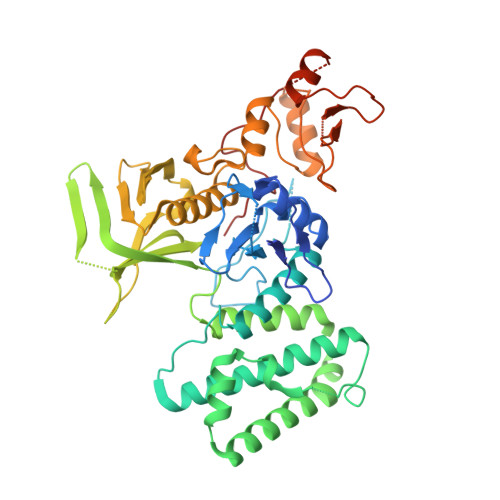
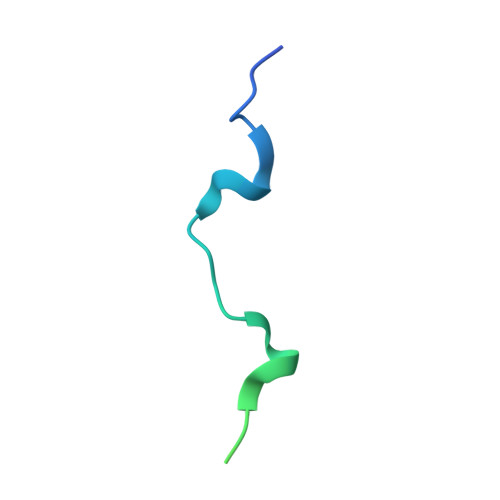
The Structural Basis for Class II Cytokine Receptor Recognition by JAK1.
Ferrao, R., Wallweber, H.J., Ho, H., Tam, C., Franke, Y., Quinn, J., Lupardus, P.J.(2016) Structure 24: 897-905
- PubMed: 27133025
- DOI: https://doi.org/10.1016/j.str.2016.03.023
- Primary Citation of Related Structures:
5IXD, 5IXI - PubMed Abstract:
JAK1 is a member of the Janus kinase (JAK) family of non-receptor tyrosine kinases that are activated in response to cytokines and interferons. Here, we present two crystal structures of the human JAK1 FERM and SH2 domains bound to peptides derived from the class II cytokine receptors IFN-λ receptor 1 and IL-10 receptor 1 (IFNLR1 and IL10RA). These structures reveal an interaction site in the JAK1 FERM that accommodates the so-called "box1" membrane-proximal receptor peptide motif. Biophysical analysis of the JAK1-IFNLR1 interaction indicates that the receptor box1 is the primary driver of the JAK1 interaction, and identifies residues conserved among class II receptors as important for binding. In addition, we demonstrate that a second "box2" receptor motif further stabilizes the JAK1-IFNLR1 complex. Together, these data identify a conserved JAK binding site for receptor peptides and elucidate the mechanism by which class II cytokine receptors interact with JAK1.
- Department of Structural Biology, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: