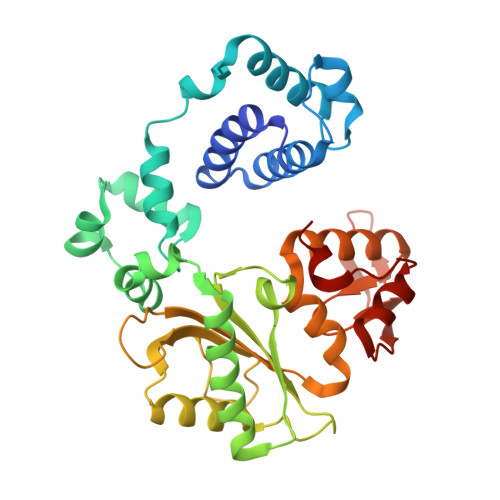
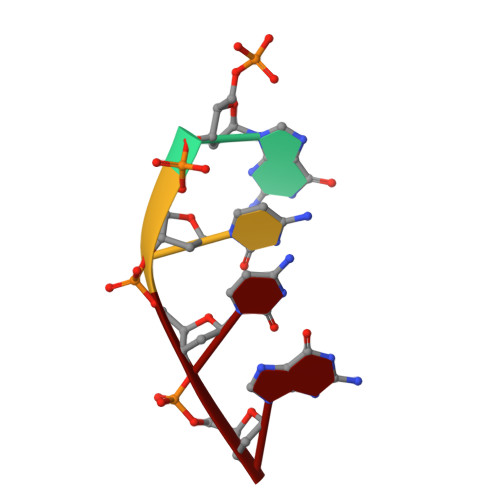
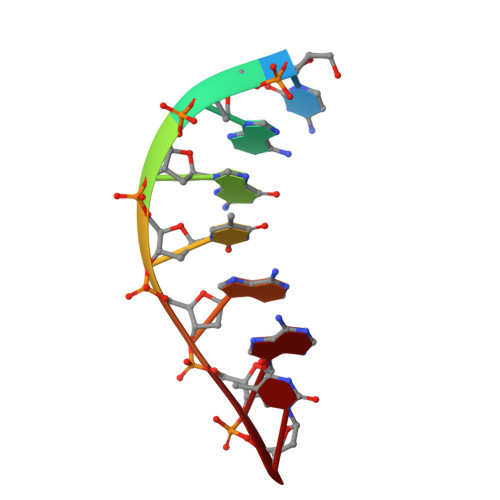
A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Burak, M.J., Guja, K.E., Hambardjieva, E., Derkunt, B., Garcia-Diaz, M.(2016) EMBO J 35: 2045-2059
- PubMed: 27481934
- DOI: https://doi.org/10.15252/embj.201694332
- Primary Citation of Related Structures:
5III, 5IIJ, 5IIK, 5IIL, 5IIM, 5IIN, 5IIO - PubMed Abstract:
8-oxo-7,8-dihydroxy-2'-deoxyguanosine (8-oxo-dG) has high mutagenic potential as it is prone to mispair with deoxyadenine (dA). In order to maintain genomic integrity, post-replicative 8-oxo-dG:dA mispairs are removed through DNA polymerase lambda (Pol λ)-dependent MUTYH-initiated base excision repair (BER). Here, we describe seven novel crystal structures and kinetic data that fully characterize 8-oxo-dG bypass by Pol λ. We demonstrate that Pol λ has a flexible active site that can tolerate 8-oxo-dG in either the anti- or syn-conformation. Importantly, we show that discrimination against the pro-mutagenic syn-conformation occurs at the extension step and identify the residue responsible for this selectivity. This residue acts as a kinetic switch, shunting repair toward long-patch BER upon correct dCMP incorporation, thus enhancing repair efficiency. Moreover, this switch also provides a potential mechanism to increase repair fidelity of MUTYH-initiated BER.
- Department of Pharmacological Sciences, Stony Brook University, Stony Brook, NY, USA.
Organizational Affiliation: