The crystal structure of SdrE from staphylococcus aureus
Zhang, S., Wei, J., Wu, S., Zhang, X., Luo, M., Wang, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine-aspartate repeat-containing protein E | 314 | Staphylococcus aureus subsp. aureus MSSA476 | Mutation(s): 0 Gene Names: sdrE, SAS0521 | 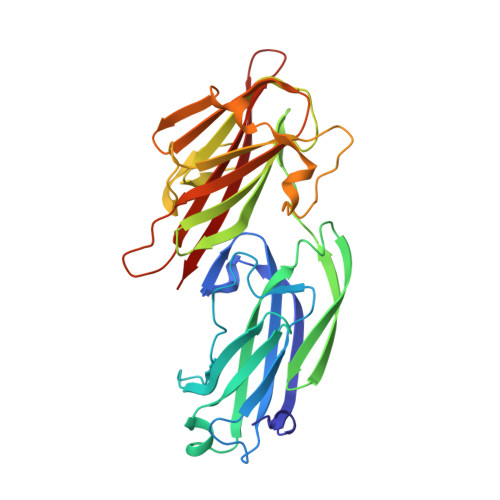 | |
UniProt | |||||
Find proteins for Q6GBS4 (Staphylococcus aureus (strain MSSA476)) Explore Q6GBS4 Go to UniProtKB: Q6GBS4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6GBS4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 39.26 | α = 95.59 |
| b = 41.56 | β = 97.55 |
| c = 51.14 | γ = 99.55 |
| Software Name | Purpose |
|---|---|
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| iMOSFLM | data reduction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China | China | 81301395 |
| Chongqing Natural Science Foundation | China | cstc2015jcyjA10003 |