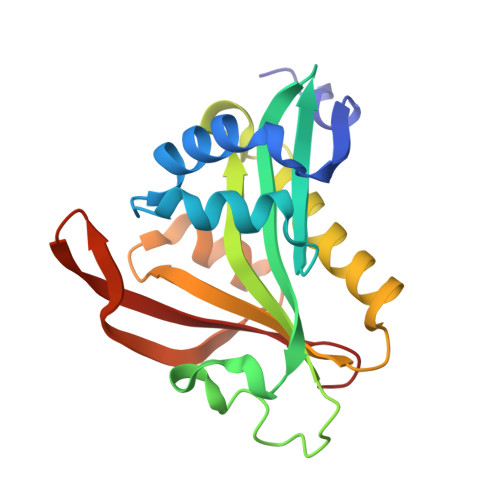
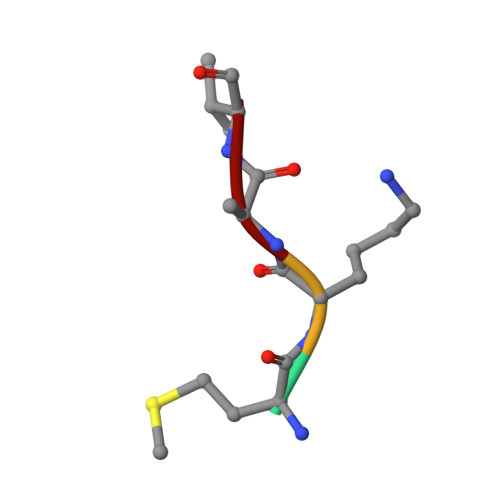
Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Stve, S.I., Magin, R.S., Foyn, H., Haug, B.E., Marmorstein, R., Arnesen, T.(2016) Structure 24: 1044-1056
- PubMed: 27320834
- DOI: https://doi.org/10.1016/j.str.2016.04.020
- Primary Citation of Related Structures:
5ICV, 5ICW - PubMed Abstract:
N-Terminal acetylation is a common and important protein modification catalyzed by N-terminal acetyltransferases (NATs). Six human NATs (NatA-NatF) contain one catalytic subunit each, Naa10 to Naa60, respectively. In contrast to the ribosome-associated NatA to NatE, NatF/Naa60 specifically associates with Golgi membranes and acetylates transmembrane proteins. To gain insight into the molecular basis for the function of Naa60, we developed an Naa60 bisubstrate CoA-peptide conjugate inhibitor, determined its X-ray structure when bound to CoA and inhibitor, and carried out biochemical experiments. We show that Naa60 adapts an overall fold similar to that of the catalytic subunits of ribosome-associated NATs, but with the addition of two novel elongated loops that play important roles in substrate-specific binding. One of these loops mediates a dimer to monomer transition upon substrate-specific binding. Naa60 employs a catalytic mechanism most similar to Naa50. Collectively, these data reveal the molecular basis for Naa60-specific acetyltransferase activity with implications for its Golgi-specific functions.
- Department of Molecular Biology, University of Bergen, 5020 Bergen, Norway; Department of Surgery, Haukeland University Hospital, 5021 Bergen, Norway.
Organizational Affiliation: