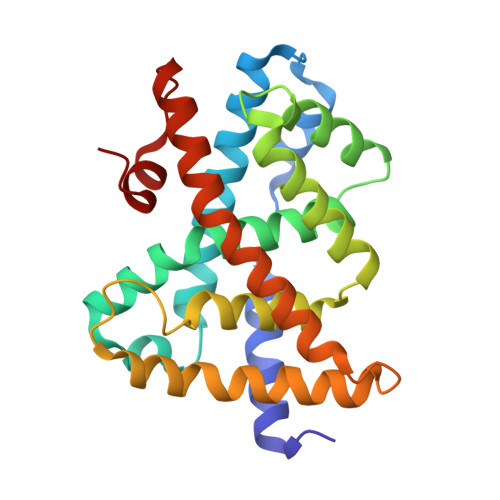
A Novel Class of Natural FXR Modulators with a Unique Mode of Selective Co-regulator Assembly
Zheng, W., Lu, Y., Lin, S., Wang, R., Qiu, L., Zhu, Y., Yao, B., Guo, F., Jin, S., Jin, L., Li, Y.(2017) Chembiochem 18: 721-725
- PubMed: 28186695
- DOI: https://doi.org/10.1002/cbic.201700059
- Primary Citation of Related Structures:
5IAW, 5ICK - PubMed Abstract:
The farnesoid X receptor (FXR) is an important target for drug discovery. Small molecules induce a conformational change in FXR that modulates its binding to co-regulators, thus resulting in distinct FXR functional profiles. However, the mechanisms for selectively recruiting co-regulators by FXR remain elusive, partly because of the lack of FXR-selective modulators. We report the identification of two natural terpenoids, tschimgine and feroline, as novel FXR modulators. Remarkably, their crystal structures uncovered a secondary binding pocket important for ligand binding. Further, tschimgine or feroline induced dynamic conformational changes in the activation function 2 (AF-2) surface, thus leading to differential co-regulator recruiting profiles, modulated by both hydrophobic and selective hydrogen-bond interactions unique to specific co-regulators. Our findings thus provide a novel structure template for optimization for FXR-selective modulators of clinical value.
- State Key Laboratory of Cellular Stress Biology, School of Life Sciences, Innovation Center for Cell Signaling Network, Xiamen University, Xiamen, Fujian, 361005, China.
Organizational Affiliation: