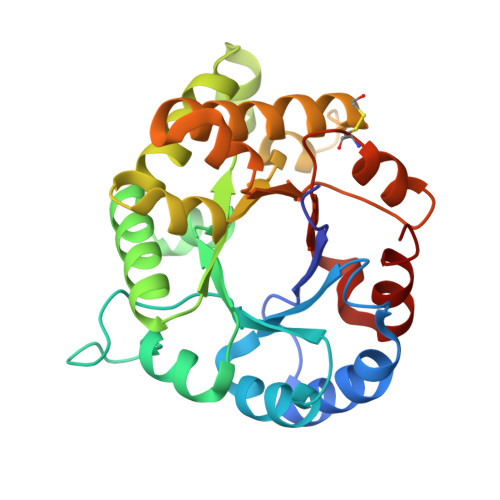
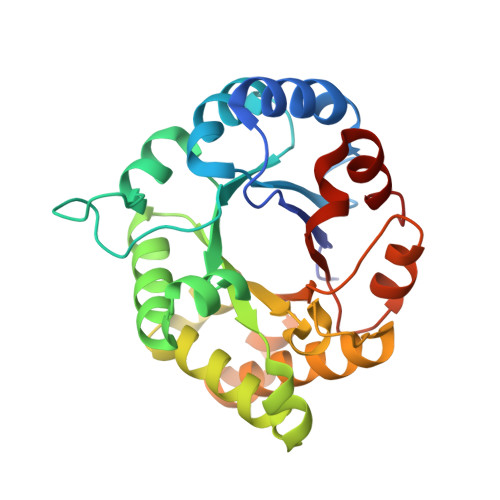
1.65 Angstrom Crystal Structure of Triosephosphate Isomerase (TIM) from Streptococcus pneumoniae
Minasov, G., Shuvalova, L., Dubrovska, I., Flores, K., Shatsman, S., Kwon, K., Anderson, W.F., Center for Structural Genomics of Infectious DiseasesTo be published.