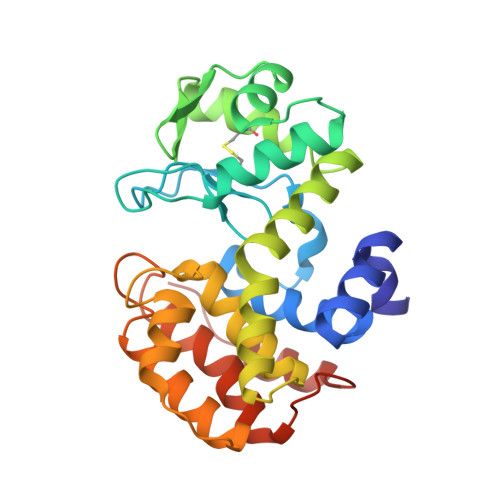
Crystal structure of the GH-46 subclass III chitosanase from Bacillus circulans MH-K1 in complex with chitotetraose
Suzuki, M., Saito, A., Kobayashi, M., Yokoyama, T., Omiya, S., Li, J., Sugita, K., Miki, K., Saito, J.I., Ando, A.(2024) Biomed Biochim Acta 1868: 130549