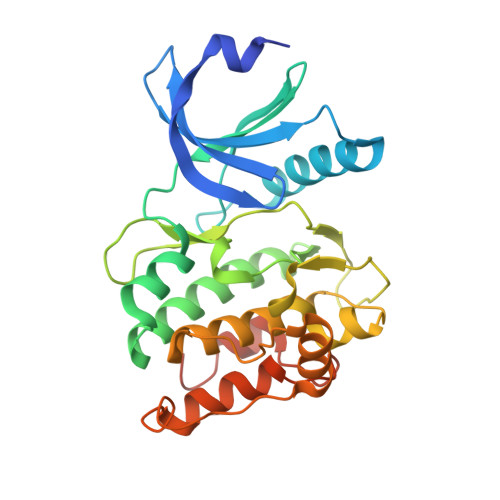
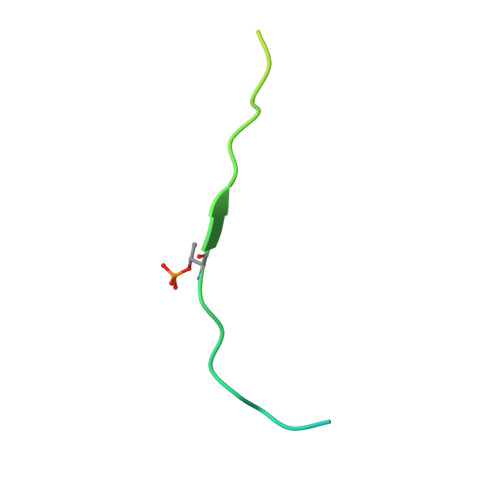
The Interaction between the Drosophila EAG Potassium Channel and the Protein Kinase CaMKII Involves an Extensive Interface at the Active Site of the Kinase.
Castro-Rodrigues, A.F., Zhao, Y., Fonseca, F., Gabant, G., Cadene, M., Robertson, G.A., Morais-Cabral, J.H.(2018) J Mol Biology 430: 5029-5049
- PubMed: 30381148
- DOI: https://doi.org/10.1016/j.jmb.2018.10.015
- Primary Citation of Related Structures:
5FG8, 5H9B, 5HU3 - PubMed Abstract:
The Drosophila EAG (dEAG) potassium channel is the founding member of the superfamily of KNCH channels, which are involved in cardiac repolarization, neuronal excitability and cellular proliferation. In flies, dEAG is involved in regulation of neuron firing and assembles with CaMKII to form a complex implicated in memory formation. We have characterized the interaction between the kinase domain of CaMKII and a 53-residue fragment of the dEAG channel that includes a canonical CaMKII recognition sequence. Crystal structures together with biochemical/biophysical analysis show a substrate-kinase complex with an unusually tight and extensive interface that appears to be strengthened by phosphorylation of the channel fragment. Electrophysiological recordings show that catalytically active CaMKII is required to observe active dEAG channels. A previously identified phosphorylation site in the recognition sequence is not the substrate for this crucial kinase activity, but rather contributes importantly to the tight interaction of the kinase with the channel. The available data suggest that the dEAG channel is a docking platform for the kinase and that phosphorylation of the channel's kinase recognition sequence modulates the strength of the interaction between the channel and the kinase.
- IBMC, Instituto de Biologia Molecular e Celular, Universidade do Porto, Porto, Portugal; i3S, Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Porto, Portugal.
Organizational Affiliation: