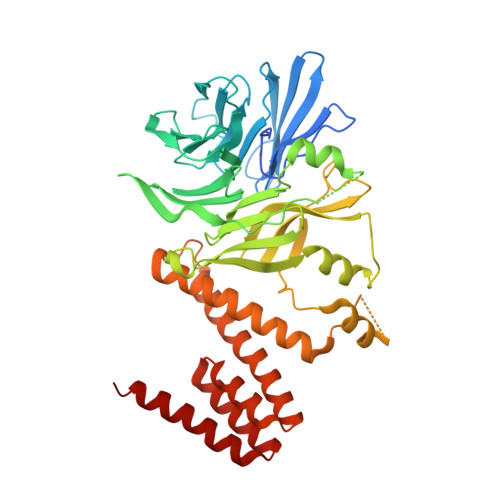
Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase.
Villa, F., Simon, A.C., Ortiz Bazan, M.A., Kilkenny, M.L., Wirthensohn, D., Wightman, M., Matak-Vinkovic, D., Pellegrini, L., Labib, K.(2016) Mol Cell 63: 385-396
- PubMed: 27397685
- DOI: https://doi.org/10.1016/j.molcel.2016.06.009
- Primary Citation of Related Structures:
5HOG, 5HOI - PubMed Abstract:
Replisome assembly at eukaryotic replication forks connects the DNA helicase to DNA polymerases and many other factors. The helicase binds the leading-strand polymerase directly, but is connected to the Pol α lagging-strand polymerase by the trimeric adaptor Ctf4. Here, we identify new Ctf4 partners in addition to Pol α and helicase, all of which contain a "Ctf4-interacting-peptide" or CIP-box. Crystallographic analysis classifies CIP-boxes into two related groups that target different sites on Ctf4. Mutations in the CIP-box motifs of the Dna2 nuclease or the rDNA-associated protein Tof2 do not perturb DNA synthesis genome-wide, but instead lead to a dramatic shortening of chromosome 12 that contains the large array of rDNA repeats. Our data reveal unexpected complexity of Ctf4 function, as a hub that connects multiple accessory factors to the replisome. Most strikingly, Ctf4-dependent recruitment of CIP-box proteins couples other processes to DNA synthesis, including rDNA copy-number regulation.
- MRC Protein Phosphorylation and Ubiquitylation Unit, Sir James Black Centre, School of Life Sciences, University of Dundee, Dow Street, Dundee DD1 5EH, UK.
Organizational Affiliation: