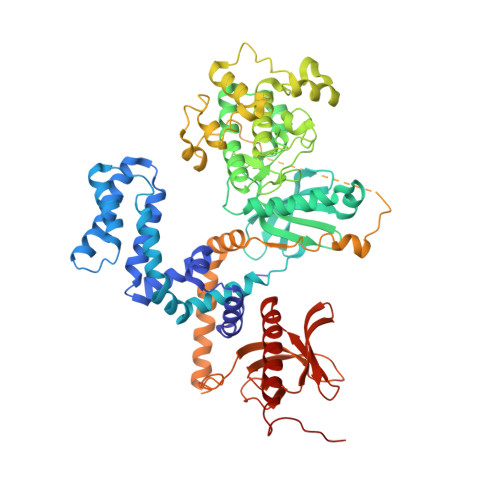
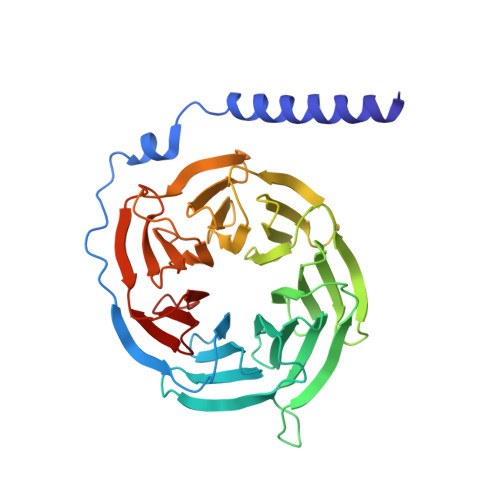
Structure-Based Design, Synthesis, and Biological Evaluation of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors.
Waldschmidt, H.V., Homan, K.T., Cruz-Rodriguez, O., Cato, M.C., Waninger-Saroni, J., Larimore, K.M., Cannavo, A., Song, J., Cheung, J.Y., Kirchhoff, P.D., Koch, W.J., Tesmer, J.J., Larsen, S.D.(2016) J Med Chem 59: 3793-3807
- PubMed: 27050625
- DOI: https://doi.org/10.1021/acs.jmedchem.5b02000
- Primary Citation of Related Structures:
5HE0, 5HE1, 5HE2, 5HE3 - PubMed Abstract:
G protein-coupled receptors (GPCRs) are central to many physiological processes. Regulation of this superfamily of receptors is controlled by GPCR kinases (GRKs), some of which have been implicated in heart failure. GSK180736A, developed as a Rho-associated coiled-coil kinase 1 (ROCK1) inhibitor, was identified as an inhibitor of GRK2 and co-crystallized in the active site. Guided by its binding pose overlaid with the binding pose of a known potent GRK2 inhibitor, Takeda103A, a library of hybrid inhibitors was developed. This campaign produced several compounds possessing high potency and selectivity for GRK2 over other GRK subfamilies, PKA, and ROCK1. The most selective compound, 12n (CCG-224406), had an IC50 for GRK2 of 130 nM, >700-fold selectivity over other GRK subfamilies, and no detectable inhibition of ROCK1. Four of the new inhibitors were crystallized with GRK2 to give molecular insights into the binding and kinase selectivity of this class of inhibitors.
- Center for Translational Medicine, Temple University , Philadelphia, Pennsylvania 19140, United States.
Organizational Affiliation: