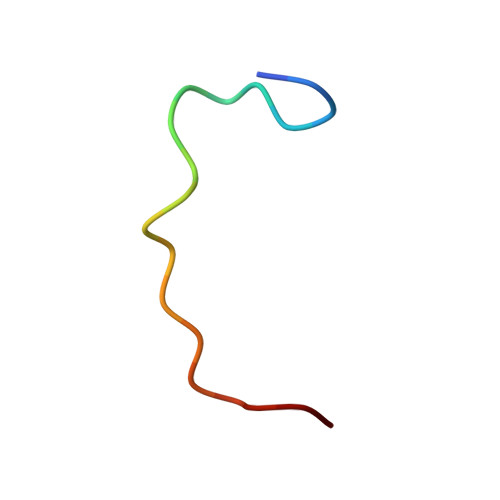
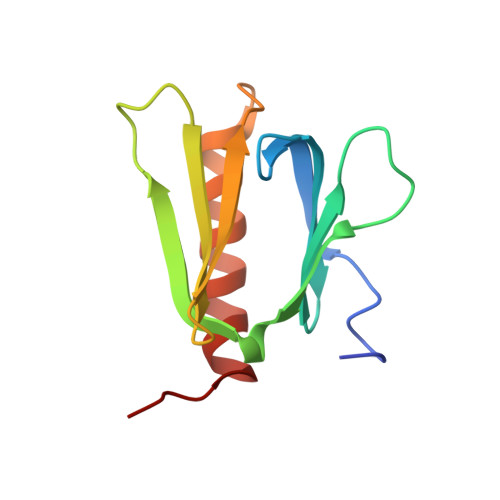
The Interaction Mode of the Acidic Region of the Cell Cycle Transcription Factor DP1 with TFIIH
Okuda, M., Araki, K., Ohtani, K., Nishimura, Y.(2016) J Mol Biology 428: 4993-5006
- PubMed: 27825926
- DOI: https://doi.org/10.1016/j.jmb.2016.11.001
- Primary Citation of Related Structures:
5GOW - PubMed Abstract:
The heterodimeric transcription factor E2F1-DP1 plays crucial roles in coordinating gene expression during G 1 /S cell cycle progression. For transcriptional activation, the transactivation domain (TAD) of E2F1 is known to interact with the TATA-binding protein of TFIID and the p62 subunit of TFIIH. It is generally believed that DP1 facilitates E2F1 binding to target DNA and does not possess a TAD. Here, we show that an acidic region of DP1, whose function has remained elusive, binds to the plekstrin homology (PH) domain of p62 with higher affinity than that of E2F1 and contributes to transcriptional activation. The structure of the complex revealed that DP1 forms a twisted U-shaped, string-like conformation and binds to the surface of the PH domain by anchoring Phe403 into a pocket in the PH domain. The transcriptional activity of E2F1-DP1 was reduced when Phe403 of DP1 was mutated. These findings indicate that the acidic region of DP1 acts as a TAD by contacting TFIIH.
- Graduate School of Medical Life Science, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.
Organizational Affiliation: