Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis.
Luna-Zurita, L., Stirnimann, C.U., Glatt, S., Kaynak, B.L., Thomas, S., Baudin, F., Samee, M.A.H., He, D., Small, E.M., Mileikovsky, M., Nagy, A., Holloway, A.K., Pollard, K.S., Muller, C.W., Bruneau, B.G.(2016) Cell 164: 999
- PubMed: 26875865
- DOI: https://doi.org/10.1016/j.cell.2016.01.004
- Primary Citation of Related Structures:
5FLV - PubMed Abstract:
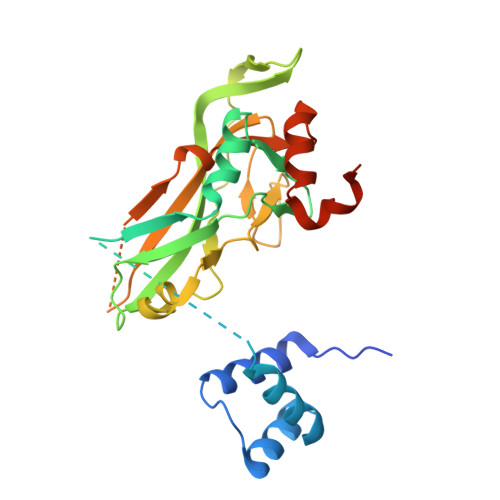
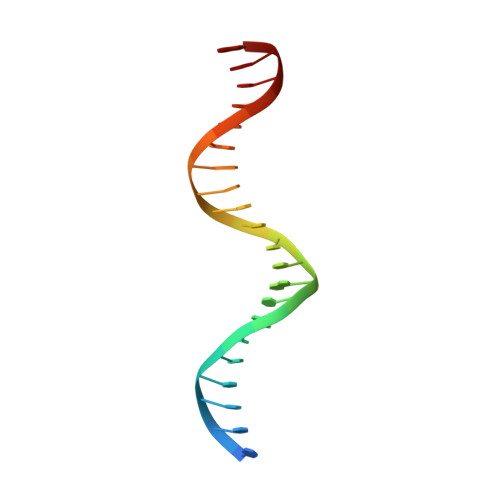
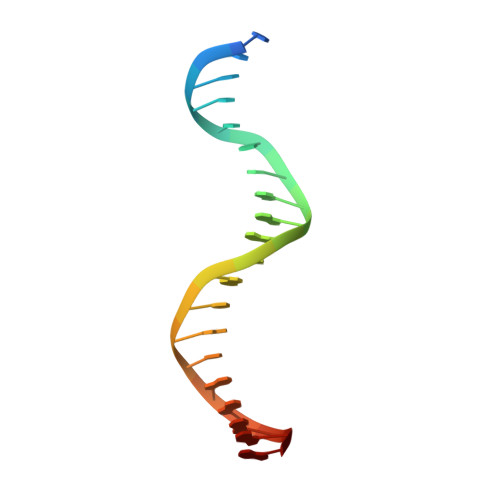
Transcription factors (TFs) are thought to function with partners to achieve specificity and precise quantitative outputs. In the developing heart, heterotypic TF interactions, such as between the T-box TF TBX5 and the homeodomain TF NKX2-5, have been proposed as a mechanism for human congenital heart defects. We report extensive and complex interdependent genomic occupancy of TBX5, NKX2-5, and the zinc finger TF GATA4 coordinately controlling cardiac gene expression, differentiation, and morphogenesis. Interdependent binding serves not only to co-regulate gene expression but also to prevent TFs from distributing to ectopic loci and activate lineage-inappropriate genes. We define preferential motif arrangements for TBX5 and NKX2-5 cooperative binding sites, supported at the atomic level by their co-crystal structure bound to DNA, revealing a direct interaction between the two factors and induced DNA bending. Complex interdependent binding mechanisms reveal tightly regulated TF genomic distribution and define a combinatorial logic for heterotypic TF regulation of differentiation.
- Gladstone Institute of Cardiovascular Disease, San Francisco, CA 94158, USA; Roddenberry Center for Stem Cell Biology and Medicine at Gladstone, San Francisco, CA 94158, USA.
Organizational Affiliation: