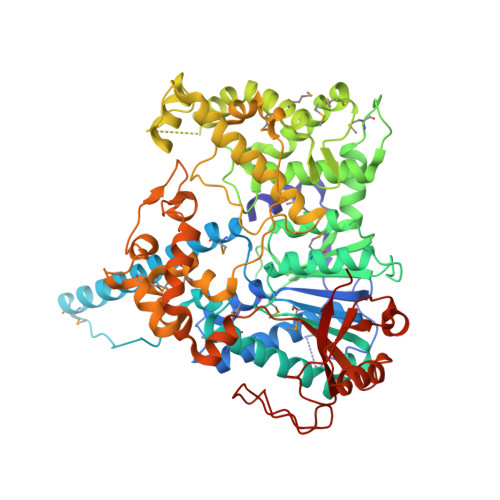
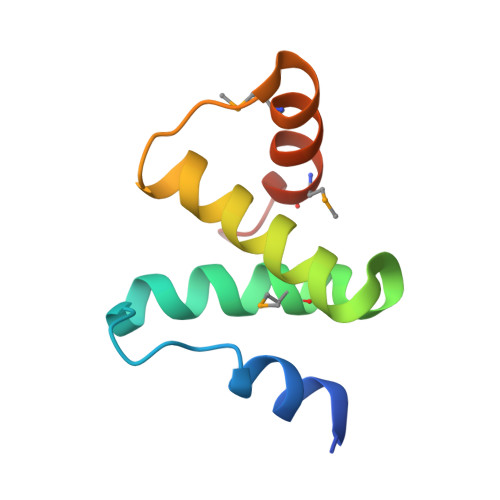
Structural Basis and Function of Xrn2-Binding by Xtb Domains
Richter, H., Katic, I., Gut, H., Grosshans, H.(2016) Nat Struct Mol Biol 23: 164
- PubMed: 26779609
- DOI: https://doi.org/10.1038/nsmb.3155
- Primary Citation of Related Structures:
5FIR - PubMed Abstract:
The RNase XRN2 is essential in RNA metabolism. In Caenorhabditis elegans, XRN2 functions with PAXT-1, which shares a putative XRN2-binding domain (XTBD) with otherwise unrelated mammalian proteins. Here, we characterize the structure and function of an XTBD-XRN2 complex. Although XTBD stably interconnects two XRN2 domains through numerous interacting residues, mutation of a single critical residue suffices to disrupt XTBD-XRN2 complexes in vitro and to recapitulate paxt-1-null mutant phenotypes in vivo. Demonstrating conservation of function, vertebrate XTBD-containing proteins bind XRN2 in vitro, and human CDKN2AIPNL (HsC2AIL) can substitute for PAXT-1 in vivo. In vertebrates, which express three distinct XTBD-containing proteins, XRN2 may partition into distinct stable heterodimeric complexes, which probably differ in subcellular localization or function. In C. elegans, complex formation with PAXT-1, the sole XTBD protein, serves to preserve the stability of XRN2 in the absence of substrate.
- Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
Organizational Affiliation: