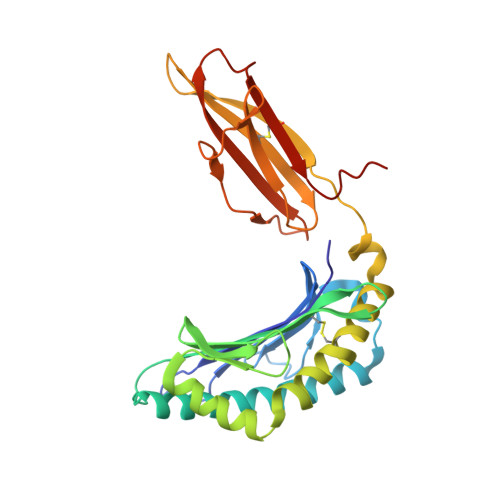
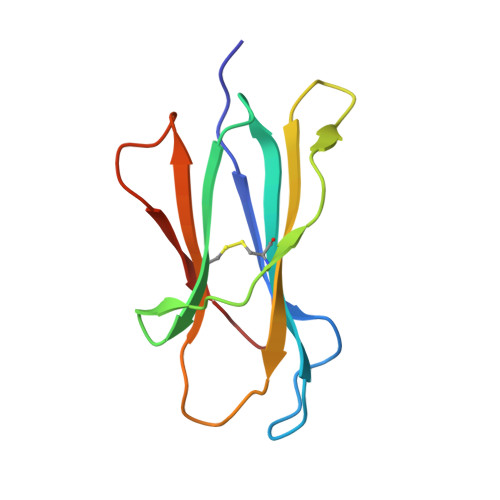
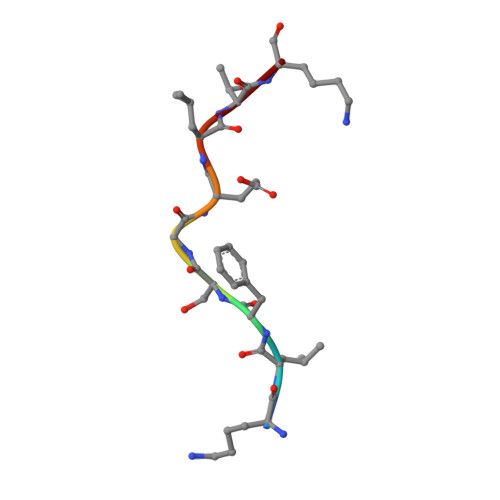
Diversified Anchoring Features the Peptide Presentation of DLA-88*50801: First Structural Insight into Domestic Dog MHC Class I
Xiao, J., Xiang, W., Chai, Y., Haywood, J., Qi, J., Ba, L., Qi, P., Wang, M., Liu, J., Gao, G.F.(2016) J Immunol 197: 2306-2315
- PubMed: 27511732
- DOI: https://doi.org/10.4049/jimmunol.1600887
- Primary Citation of Related Structures:
5F1I, 5F1N - PubMed Abstract:
Canines represent a crucial animal model for studying human diseases and organ transplantation, as well as the evolution of domestic animals. MHCs, with a central role in cellular immunity, are commonly used in the study of dog population genetics and genome evolution. However, the molecular basis for the peptide presentation of dog MHC remains largely unknown. In this study, peptide presentation by canine MHC class I DLA-88*50801 was structurally determined, revealing diversified anchoring modes of the binding peptides. Flexible and large pockets composed of both hydrophobic and hydrophilic residues can accommodate pathogen-derived peptides with diverse anchor residues, as confirmed by thermostability measurements. Furthermore, DLA-88*50801 contains an unusual α2 helix with a large coil in the TCR contact region. These results further our understanding of canine T cell immunity through peptide presentation of MHC class I and shed light on the molecular basis for vaccine development for canine infectious diseases, for example, canine distemper virus.
- Key Laboratory of Veterinary Bioproduction and Chemical Medicine of the Ministry of Agriculture, Zhongmu Institutes of China Animal Husbandry Industry Co. Ltd, Beijing 100095, China; College of Veterinary Medicine, China Agricultural University, Beijing 100193, China; China Research Network of Immunity and Health, Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing 100101, China;
Organizational Affiliation: