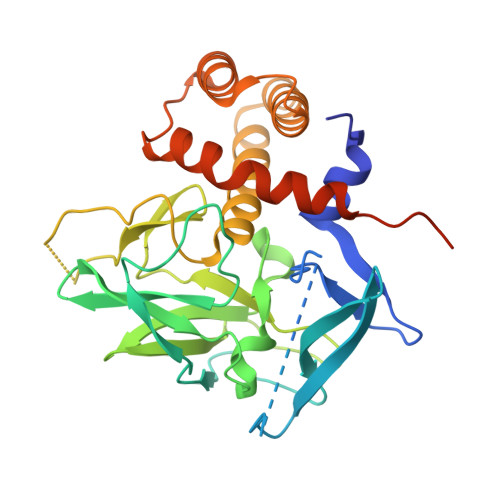
ETO family protein Mtgr1 mediates Prdm14 functions in stem cell maintenance and primordial germ cell formation.
Nady, N., Gupta, A., Ma, Z., Swigut, T., Koide, A., Koide, S., Wysocka, J.(2015) Elife 4: e10150-e10150
- PubMed: 26523391
- DOI: https://doi.org/10.7554/eLife.10150
- Primary Citation of Related Structures:
5ECJ - PubMed Abstract:
Prdm14 is a sequence-specific transcriptional regulator of embryonic stem cell (ESC) pluripotency and primordial germ cell (PGC) formation. It exerts its function, at least in part, through repressing genes associated with epigenetic modification and cell differentiation. Here, we show that this repressive function is mediated through an ETO-family co-repressor Mtgr1, which tightly binds to the pre-SET/SET domains of Prdm14 and co-occupies its genomic targets in mouse ESCs. We generated two monobodies, synthetic binding proteins, targeting the Prdm14 SET domain and demonstrate their utility, respectively, in facilitating crystallization and structure determination of the Prdm14-Mtgr1 complex, or as genetically encoded inhibitor of the Prdm14-Mtgr1 interaction. Structure-guided point mutants and the monobody abrogated the Prdm14-Mtgr1 association and disrupted Prdm14's function in mESC gene expression and PGC formation in vitro. Altogether, our work uncovers the molecular mechanism underlying Prdm14-mediated repression and provides renewable reagents for studying and controlling Prdm14 functions.
- Department of Chemical and Systems Biology, Stanford University School of Medicine, Stanford, United States.
Organizational Affiliation: