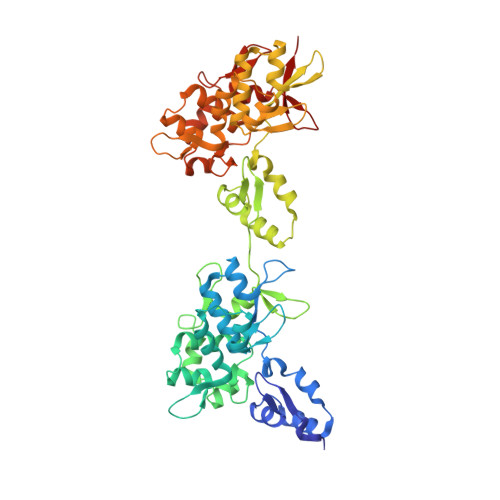Structure of the Intermediate Filament-Binding Region of Desmoplakin.
Kang, H., Weiss, T.M., Bang, I., Weis, W.I., Choi, H.J.(2016) PLoS One 11: e0147641-e0147641
- PubMed: 26808545
- DOI: https://doi.org/10.1371/journal.pone.0147641
- Primary Citation of Related Structures:
5DZZ - PubMed Abstract:
Desmoplakin (DP) is a cytoskeletal linker protein that connects the desmosomal cadherin/plakoglobin/plakophilin complex to intermediate filaments (IFs). The C-terminal region of DP (DPCT) mediates IF binding, and contains three plakin repeat domains (PRDs), termed PRD-A, PRD-B and PRD-C. Previous crystal structures of PRDs B and C revealed that each is formed by 4.5 copies of a plakin repeat (PR) and has a conserved positively charged groove on its surface. Although PRDs A and B are linked by just four amino acids, B and C are separated by a 154 residue flexible linker, which has hindered crystallographic analysis of the full DPCT. Here we present the crystal structure of a DPCT fragment spanning PRDs A and B, and elucidate the overall architecture of DPCT by small angle X-ray scattering (SAXS) analysis. The structure of PRD-A is similar to that of PRD-B, and the two domains are arranged in a quasi-linear arrangement, and separated by a 4 amino acid linker. Analysis of the B-C linker region using secondary structure prediction and the crystal structure of a homologous linker from the cytolinker periplakin suggests that the N-terminal ~100 amino acids of the linker form two PR-like motifs. SAXS analysis of DPCT indicates an elongated but non-linear shape with Rg = 51.5 Å and Dmax = 178 Å. These data provide the first structural insights into an IF binding protein containing multiple PRDs and provide a foundation for studying the molecular basis of DP-IF interactions.
- Dept. of Biological Sciences, Seoul National University, Seoul, South Korea.
Organizational Affiliation: