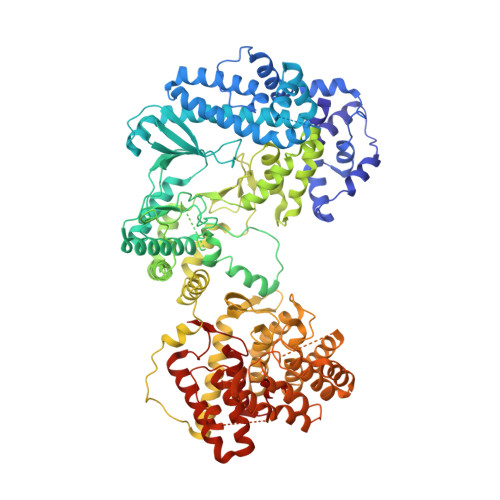
The enterococcal cytolysin synthetase has an unanticipated lipid kinase fold.
Dong, S.H., Tang, W., Lukk, T., Yu, Y., Nair, S.K., van der Donk, W.A.(2015) Elife 4
- PubMed: 26226635
- DOI: https://doi.org/10.7554/eLife.07607
- Primary Citation of Related Structures:
5DZT - PubMed Abstract:
The enterococcal cytolysin is a virulence factor consisting of two post-translationally modified peptides that synergistically kill human immune cells. Both peptides are made by CylM, a member of the LanM lanthipeptide synthetases. CylM catalyzes seven dehydrations of Ser and Thr residues and three cyclization reactions during the biosynthesis of the cytolysin large subunit. We present here the 2.2 Å resolution structure of CylM, the first structural information on a LanM. Unexpectedly, the structure reveals that the dehydratase domain of CylM resembles the catalytic core of eukaryotic lipid kinases, despite the absence of clear sequence homology. The kinase and phosphate elimination active sites that affect net dehydration are immediately adjacent to each other. Characterization of mutants provided insights into the mechanism of the dehydration process. The structure is also of interest because of the interactions of human homologs of lanthipeptide cyclases with kinases such as mammalian target of rapamycin.
- Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, United States.
Organizational Affiliation: